Thank you to everyone who joined our webcast, “Whole Genome Trait Association in SVS.” If you missed the live event and are interested in knowing what we talked about, you may access the recorded event below:
Our Live Q&A generated a lot of great questions. Unfortunately, we were unable to answer them all, but we have compiled some of the great questions our audience asked.
“Please elaborate more about GBLUP and how you would handle the analysis looking to study the evolution of genomic pattern related to various environmental and psychological factors”
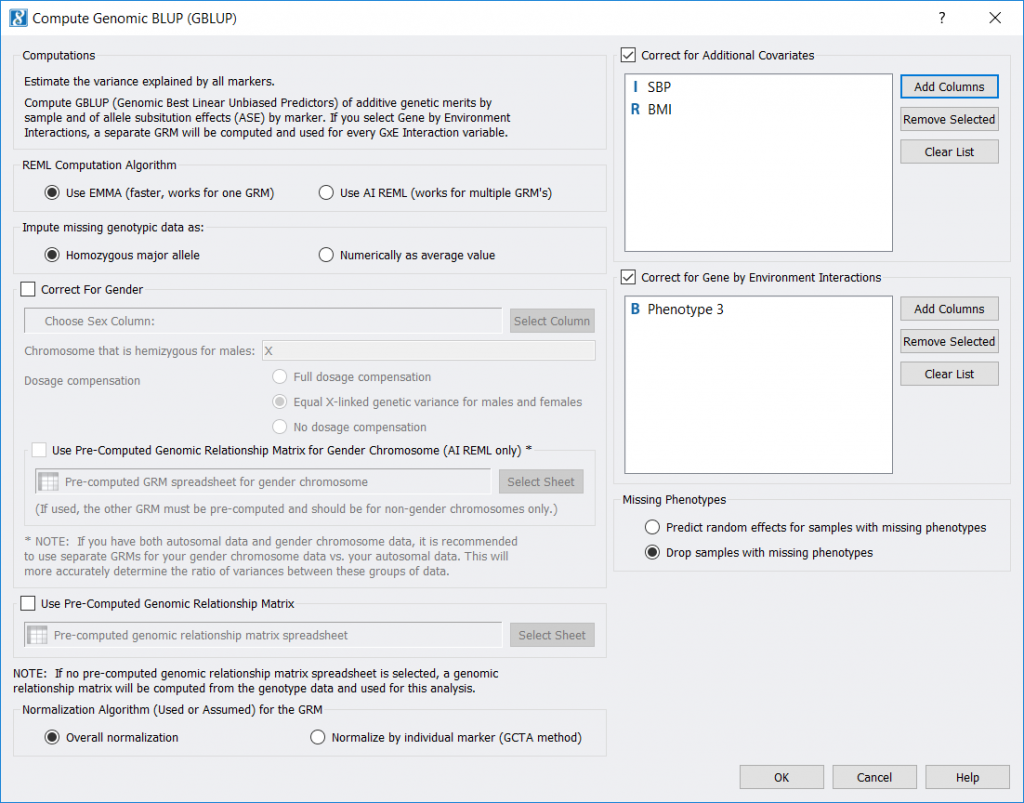
You can add covariates that represent environmental or psychological factors into a GBLUP test run in SVS in order to correct for these factors. When running GBLUP, look on the right side of the GBLUP dialog to add these optional components. In the following figure, we have specified factors SBP and BMI as covariates (additional to the intercept), and we have specified factor Phenotype 3 as a Gene by Environment interaction variable.
“How is GBLUP statistically significant?”
The significance of the random effects may be tested through a likelihood-ratio test. In this test, the likelihood of the model with the random effects is compared with the likelihood of the model without the random effects. The log of the ratio of these likelihoods is then converted to a p-value. In SVS, the results of this test for any GBLUP run are shown in the Node Change Log for each final GBLUP output. You can find details in our manual under Genomic Best Linear Unbiased Predictors Analysis.
“Does copy number vary on a regular basis in humans?”
Copy-number variations (CNV) occur all the time in biological life, and like other types of variation the vast majority are common and benign. As a general rule, the larger the variant, the less likely it is to occur, so there are far fewer copy variants than small variations like SNPs and InDels. This Nature article provides a decent overview. Occasionally these copy-number variations can cause pathological conditions. This is why each of the packages in the Golden Helix software suite, including SVS, has a capability for CNV analysis that include not only copy number variant detection, but also the annotation and filtering tools to understand their potential impact in human disease.
“Bin analysis has been done in multiple sclerosis. Is any study going on in oncology?”
We are not aware of any study of multiple sclerosis (or oncology) that has used the technique of Binned GBLUP that we demonstrated. We know of one paper on multiple sclerosis which uses a different type of binning technique (simply binning by genomic location) and a different type of analysis than we have demonstrated—however, the existence of this paper illustrates the promise in general of using a marker-binning technique. As a note, SVS Binned GBLUP can take, as an input, bins defined by any binning mechanism you would like.
“Is the bin feature a plugin of the 8.8.3 or an update on a new version?”
This feature will be a part of the SVS 8.9.0 release, which will be upcoming in about the end of August.
“What is the best way to get technical help/training from your company?”
Our support team is available for web meetings as well as phone and email to help train users or resolve any issues during your analysis. You can always email (support@goldenhelix.com) to start a support session to provide technical help or training on SVS or any Golden Helix product that you have purchased or are currently evaluating.
I hope you found this to be useful in learning more about about Golden Helix’s SVS solution. This webcast is one of many SVS webcast resources we have made available for our community, you can find the others here.
If you are interested in seeing a more personalized demo for your organization, please reach out to our team and we will be more than happy to schedule a call with you. Or, if you have any additional questions not covered here, please enter them down below!