Sentieon has become well known for the dramatic improvements it supplies in speed and accuracy for secondary analysis. Luckily, these improvements are not only available for DNAseq germline based variants but also for TNSeq somatic investigation. Sentieon has proven the capability by leading in awards such as the ICGC-TCGA DREAM Challenge for SNVs, Indel, and structural variants. This value not only applies to Tumor-Normal workflows but also for Tumor-Only application.
We have put together this blog series to break down the concepts behind each workflow where there are considerations for the required files to help improve efficiency overall. The first part of the blog series focuses on a Tumor-Normal workflow (Figure 1), and the second part covers a Tumor-Only workflow (Figure 2).
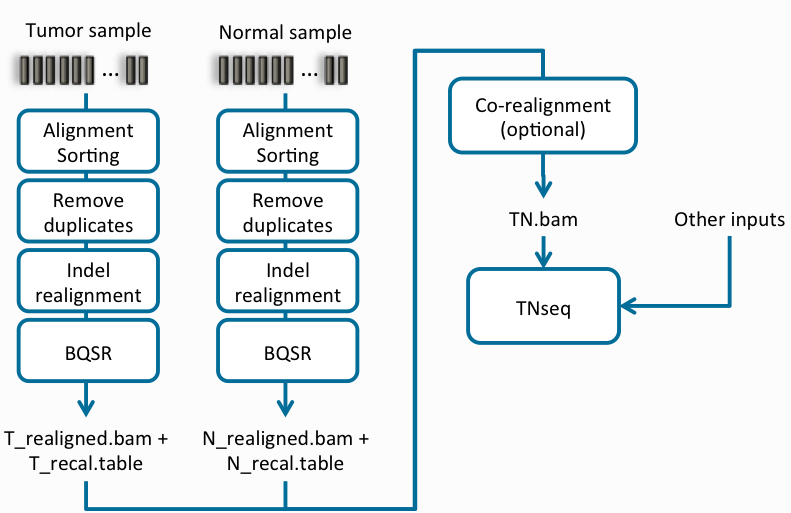

You can see in the examples above the step for removing duplicates which is an optional feature that may be deselected when running amplicon data. We also understand the need for high sensitivity when capturing somatic variants. To further amplify the potential results, Part III of this blog will cover a high sensitivity script developed to capture as many potential variants as possible.
Continue reading this series in Part II as we explore how to standardize your somatic variant calling process. As always, please reach out to support@goldenhelix.com with any questions you may have.