Here at Golden Helix, we’ve sought to develop top-quality bioinformatic software to handle not only high-throughput clinical next-gen sequencing pipelines but also cater to research groups exploring their cohorts. Our tools fit in this NGS pipeline by importing the VCF and BAM files for downstream filtration, annotation, and classification/interpretation as well as clinical reporting. It was obvious early on and even more so now that our users had a need to bring all the established secondary and tertiary tools in-house and do away with the hurdles of customized pipelines constructed from public tools on the web. These public tools are driven by innovation and global need but the reality is that these tools need optimization and ongoing support/development to keep pace with industry requirements. This was one of the motivations to partner with Sentieon advocating for a strong secondary-analysis solution to carry out the alignment and variant calling process from FASTQ to BAM/VCF. Moreover, Sentieon keeps their ear to the ground on industry needs and has since released a major update on improved calling functionality and the purpose of this blog series is to summarize these updates for our readers. Sentieon’s latest release advertises accuracy gains of 10% for Illumina based short-read approaches making this pipeline worldclass in accuracy, efforts to support new sequencing platforms include PacBio HIFI and Element Biosciences with highly accurate pipelines and more platforms available shortly as well as continuing to improve aligner efficiency for BWA-MEM, STAR, and minimap2.
Need for Speed & Long-Read:
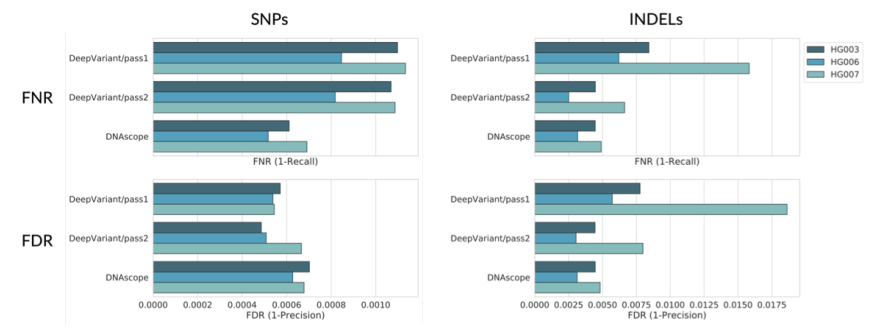
As seen on the company site, Sentieon’s focus is to develop and supply a suite of bioinformatic secondary-analysis tools that process genomic data with high computing efficiency, fast turnaround time, exceptional accuracy, and 100% consistency. Sentieon has provided major improvements in many areas of NGS short-read (150 – 300 bp) alignment and calling which began with an effort to reduce not only the run time of established alignment and calling processes with drop in replacements for BWA-MEM, GATK, and Mutect2 while also producing 100% consistent results by eliminating shortcuts like random down-sampling reads. So not only do users benefit from major speed improvements, especially relevant for exome and genome-level data but also can rely on having 100% consistency with results. The efforts did not stop with short reads and now Sentieon is providing major improvements on accuracy and speed for long read (10k – 100k bp) technology as well. A recent blog from late 2021 summarizes the results of stacking Google’s DeepVariant caller against Sentieon’s DNAscope utilizing PacBio HiFi long-read technology. A summary of results can be seen in the image below and overall Sentieon was the top performer with lower false-negative calls for snvs and indels and a respectful level of false discoveries which can be easily filtered in the tertiary stage in Golden Helix’s VarSeq software downstream.
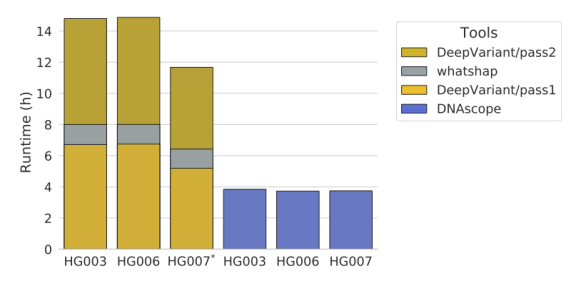
Moreover, users not only care about quality but also speed. Secondary solutions need to operate within a reasonable time frame to not only handle large datasets (exome and genome-level data) in a practical manner but also facilitate the rapid turnover needed in clinical environments. The DNAscope vs DeepVariant test was also considerate for runtime which Sentieon was the top performer with both platforms generating phased genotypes.
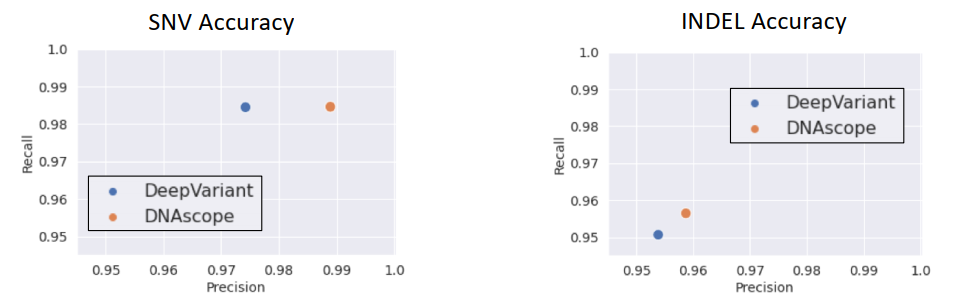
Additionally, one major hurdle with short-reads is accurate variant calling over highly repetitive regions, low-complexity regions, segmentally duplicated regions, indels spanning 15-50 bp, or small CNVs (Wagner et al. 2022). These Challenging Medically Relevant Genes or CMRGs served as a benchmark for PacBio long-read- technology. A comparison of DeepVariant against Sentieon’s DNAscope variant caller shows DNAscope in the lead for SNP and indel accuracy against the GiaB CMRG truth set.
The take-home message here is that Sentieon continues to improve their software for Illumina short reads and is supporting additional platforms beyond Illumina as they enter the market. These new improvements in accuracy and efficiency are worth checking out. Sentieon software offers highly accurate, efficient, and easy to use pipelines. We will cover some additional advancements Sentieon’s accuracy and run time in an upcoming blog. If you would like to test this product or open a discussion with the Golden Helix support team on installation/utilization please email us at support@goldenhelix.com.
Wagner, J., Olson, N.D., Harris, L. et al. Curated variation benchmarks for challenging medically relevant autosomal genes. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-021-01158-1