The collaboration between the Clinical Genome Resource (ClinGen) consortium and the American College of Medical Genetics (ACMG) recently developed published guidelines for the interpretation of CNVs called on next-generation sequencing data. These new guidelines are the first to provide a robust set of rules for the interpretation of small intragenic deletions and duplications and are now automated in VSClinical.
There is no way to hide the fact that these guidelines are extremely complex. If you deep dive into these guidelines, you will discover that there are five pages of tables describing around 80 distinct criteria. At a high level, these criteria detail different processes for scoring gains and losses and each has an associated numeric strength, both positive and negative. The summation of these values indicates evidence for, or against, pathogenicity. Fortunately, VSClinical provides a comprehensive workflow solution to guide you through these complex guidelines.
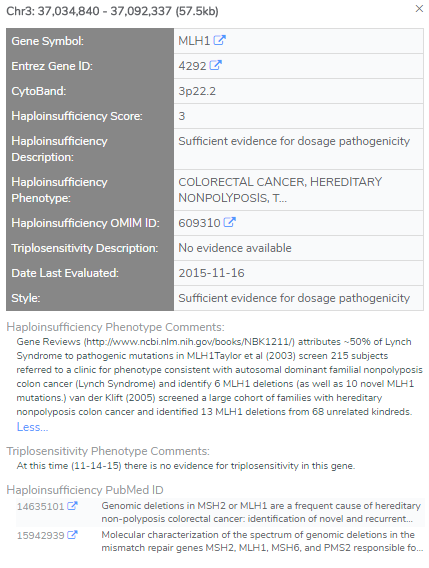
One section of the guidelines that have a strong impact on the CNV evaluation is establishing the haploinsufficiency and triplosensitivity of a given gene. This dosage sensitivity analysis must be done on a gene-by-gene basis and requires an in-depth review of the available literature associated with the given gene. Much of this work has already been done through the ClinGen Dosage Sensitivity Map, which maintains a catalog of established haploinsufficient and triplosensitive genes. As shown in Figure 1, VSClinical provides you with all relevant information and will assess if there is sufficient evidence suggesting dosage sensitivity.
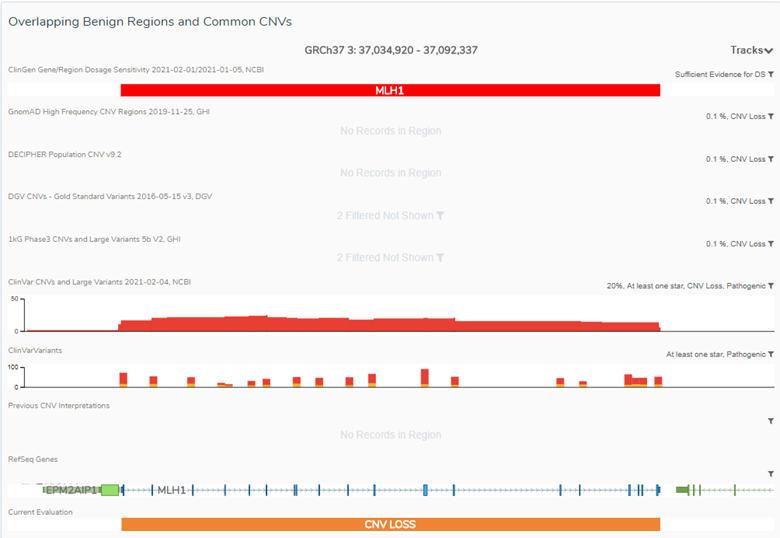
VSClinical also offers unique visualization capabilities. As shown in Figure 2, users can identify if a CNV falls within a dosage sensitivity region based on ClinGen, as well as supporting evidence of observed frequency and occurrence of CNVs within the gene or region. This includes population databases such as GnomAD High-Frequency Regions, DECIPHER, DGV CNVs, and 1KG Phase3. This interface will also provide evidence of similar events that have been observed in ClinVar CNVs and Large Variants, which can further support the pathogenicity of a given CNV. In the case of well-established genes, no additional work is needed to establish the gene’s dosage sensitivity and the clinician can limit their analysis to the CNV’s impact on the gene in question.
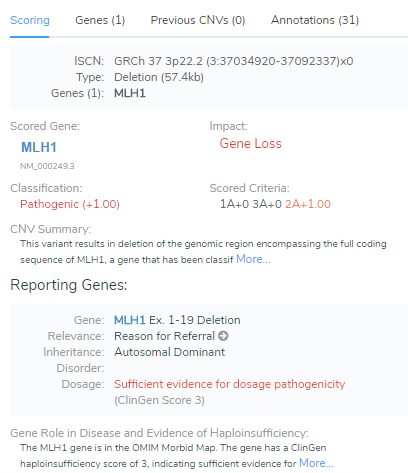
Once all of the criteria are answered, the numerical values are added together and the final classification is displayed. As shown in Figure 3, there was sufficient evidence for dosage pathogenicity which resulted in a final classification of Pathogenic. From here, users can easily incorporate the CNV Summary and the Gene Role in Disease and Evidence of Haploinsufficiency into the interpretation section, which can be easily rendered into a customizable clinical report.
Together, the ClinGen and ACMG group recently provided guidelines for the evaluation of CNVs and Golden Helix is the first in the clinical workspace to provide automation of the workflow. Here we discussed how VSClinical offers unique details and visualization capabilities for determining dosage sensitivity as well as how the information is automatically rendered. If you would like to learn more about this feature or have general questions, we would be happy to assist. All you need to do is contact support@goldenhliex.com and we will get back to you.
Thanks for reading.