Our FAS team would like to thank everyone who attended our December 2022 webcast, A User’s Perspective: Somatic Variant Analysis in VarSeq 2.3.0. This webcast allowed three members of our FAS team to give their unique insights concerning the improvements to our new VarSeq 2.3.0 release, which will be recapped here. Starting with template creation, our Technical Field Application Scientist, Solomon Reinman, gave an overview of automation capabilities with the revised VSPipeline. Next, Dr. Rana Smalling gave an overview of our Golden Helix Cancer Knowledge Base (Cancer KB), which was used to automatically render an interpretation of our somatic variant of interest. Finally, Dr. Jennifer Dankoff walked us through our newly updated AMP clinical report templates. Together, our FAS demonstrated a number of options a user could leverage to expedite their somatic analysis with VarSeq 2.3.0.
Starting from the beginning, current VarSeq users familiar with somatic workflows will still be very comfortable with template design and variant annotation and filtration in VarSeq 2.3.0. The same fundamental logic applies to annotating and filtering small variants and CNVs, though our import algorithms have been overhauled in terms of speed and versatility. On top of the standard small variant import, users can also include break-ends (fusions) and CNVs on the initial import.
On the automation side with VSPipeline, VarSeq’s command line interface, the fundamentals are also still in place. However, users now have access to much more nimble commands within VSPipeline, allowing greater depth and breadth of automation. More granular controls now allow users to switch between project states within VarSeq and access the evaluation and interpretation hubs within VSClinical in an automated fashion. Increased scripting capabilities that allow for the import of comprehensive genomic signatures can be initiated in VSPipeline, enabling users to rely on the thorough data curation efforts of our Cancer Knowledge Base team to automatically render reports encompassing a wide variety of highly vetted information.
As excited as we are about the engineering effort that has gone into providing these capabilities to our users, they would be useless without the profound curation effort that has gone into VSClinical and CancerKB. Let’s go into some more detail on the vast improvements these modules have seen in VarSeq 2.3.0.
We are delighted to announce the changes to our Golden Helix Cancer Knowledgebase. Specifically, we want to highlight Golden Helix CancerKB as the key to VSPipeline’s automation of clinical reports. Golden Helix CancerKB now automates the association of high-quality biomarker and drug interpretations with each variant that is added to VSClinical AMP, and those interpretations can therefore be pulled into a clinical report with custom scripts when running VSPipeline. This is a long-awaited feature that will see our users save time and effort as we can now enable full automation from VCF all the way to clinical reports.
Golden Helix CancerKB relies on our dedicated team of professional curators to keep our knowledge base updated monthly with the latest high-quality expert-curated interpretations for various types of cancer biomarkers and drug treatment options. Golden Helix CancerKB V2.0, which will be released with VarSeq 2.3.0 in the upcoming weeks, boasts expanded Tier 1 and 2 interpretations, including all the genes in the TSO-500 gene panel, genomic signatures such as microsatellite instability (MSI), tumor mutation burden (TMB), PD-L1 status, combination biomarkers such as KRAS and BRAF, negative findings, and gene impact biomarkers. We have revamped our drug interpretations as well to list indications, approval status, and more, and also carry drug interpretations at the level of the tumor type. Post-release, we will host a webcast to discuss the new Golden Helix CancerKB in its entirety.
Integration of these new and exciting changes to our knowledge base has allowed us to provide users with clinical reports that are in keeping with the trends in the field of clinical cancer genomics and comprehensive genomic profiling. As such, we have updated our report formats and are excited to share those with our users.
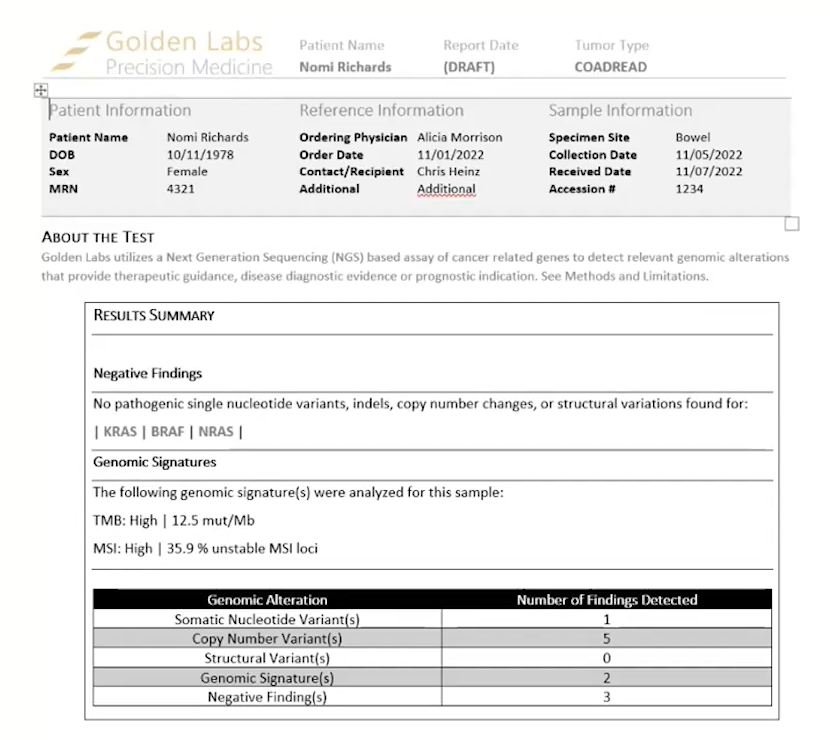
There are now three report templates available for cancer workflows. There is a Cancer Biomarkers Template, a Cancer Drugs and Trails Template V3, and an updated Cancer Report Template V3. The Cancer Biomarkers Template is new to 2.3.0 and has usual sections for variants and copy number variants, along with the newly added support for structural variants, genomic signatures, negative findings, TMB, and MSI, all which can be imported with TSO500 data (Figure 1). This template gives the detailed findings sections you would find with any Golden Helix report template.
Our other cancer report templates are very similar to their previous formats. The Cancer Drugs and Trials V3 template contains all the information fields of the previous report, with additional fields emphasizing therapeutic options, clinical trials, and drug information. The Cancer Report Template V3, like its counterparts, contains all the relevant biomarker, drug, and trial data in a very streamlined format. All of the report templates are still entirely customizable, with these templates providing solid starting points for creating a report that suits your unique workflow needs. If you would like to know more about report customization, please check out our webcast, Advanced Report Customization in VSClinical.
Let’s circle back to the topic of automation. As impressive as the expansive improvements we’ve made to the curated data available to VSClinical AMP users, one of the most exciting features in our new release is how users can capitalize on the curation effort by applying all of this knowledge in an automated fashion. As we demonstrated in our webcast, the manual steps we’ve stepped through here to generate a clinical report can be perfectly replicated in VSPipeline. Once users have a validated workflow established, it’s as simple as running a single command in the terminal to produce the same consistent results.
And with that, thank you again to those who joined us for our User Perspective webcast. We hope that this recap was informative and helpful. Many of our customers have been asking for more information about the updates to CancerKB, which Dr. Smalling briefly went over here. The good news is there will be another webcast scheduled for early Jan. 2023, which will go over, in-depth, the updates and utilities of VarSeq’s Cancer KB. Stay tuned!
Golden Helix has developed simple, fast, and repeatable variant analysis software for gene panels, exomes, and whole genomes. With an intuitive design, VarSeq allows you to create a high throughput environment by creating repeatable workflows and efficient data analysis.