Abstract
Before assessing the clinical significance of a somatic mutation, one must determine if the mutation is likely to be a driver mutation (i.e. a mutation that provides a selective growth advantage, thereby promoting cancer development). To aid clinicians in this process, VSClinical provides an oncogenicity scoring system, which uses a variety of metrics to classify a given somatic mutation into one of the following categories: oncogenic, likely oncogenic, benign, likely benign, or uncertain significance. This scoring system is heavily inspired by the ACMG Guidelines for the interpretation of germline mutations but has several important differences to make it more applicable in the context of somatic variant interpretation.
Our oncogenicity scoring system relies on an additive point system in which points are assigned to a given variant based on several criteria. The various criteria that are considered when evaluating a given variant are summarized on the table below:
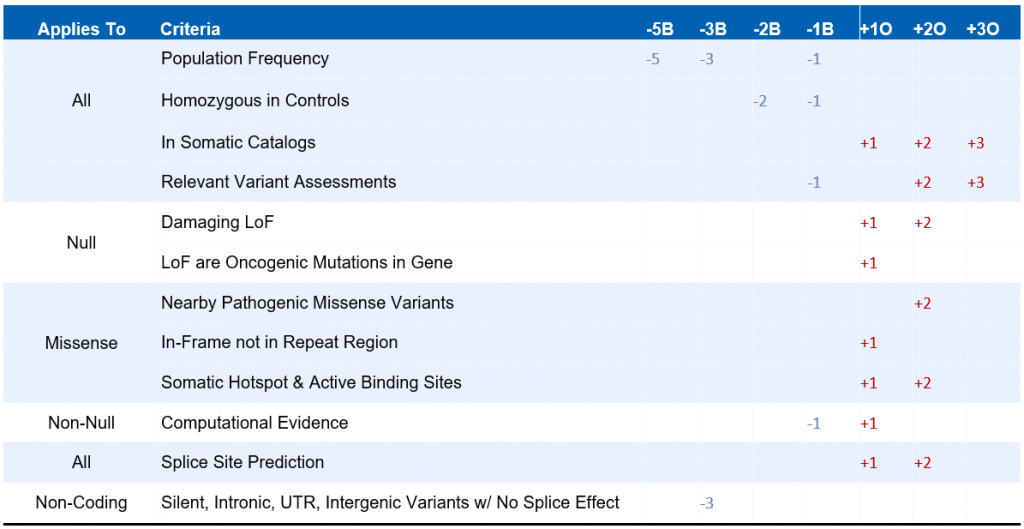
Many of the criteria above are shared by the ACMG Guidelines for germline variant interpretation, such as population frequency information, variant effect on protein function, and nearby pathogenic variants in catalogs such as ClinVar. However, other criteria are specific to the world of somatic variant interpretation. These include the variant’s presence in somatic catalogs such as COSMIC, the effect of other known oncogenic variants in the same gene, and the variant’s presence in known cancer hotspots or active binding sites. These criteria are combined by summing over the scores for all applicable scoring criteria. Scores exceeding 3 indicate an oncogenic or likely oncogenic classification, while scores falling below -3 indicate a benign or likely benign classification.
In this webcast, we discuss how each of these scoring criteria are combined to obtain an oncogenicity classification. This includes a discussion of the considerations taken into account during the development of this scoring system and a detailed analysis of several example mutations to illustrate the system in practice.
On-Demand Recording
Didn’t get a chance to join us live? Watch the on-demand recording here!
Webcast Q&A
This webcast generated several excellent questions that we would like to feature on the blog for others who may have similar questions.
Q: For the SULTIC3 W36* mutation, a frameshift mutation we covered in the video, it’s near the start of the exon of the gene. Does it not mean that the protein will not be expressed? So, why is it not pathogenic, but benign?
This is an excellent question and best answered by looking at VSClinical. Here is a short snippet from our webcast where I answer this:
Q: What is the specific evidence for excluding LoFs near the end of the gene? How are those cut points selected?
A: This is an example where we actually borrowed very heavily from the ACMG guidelines. We actually selected those cut points directly from what’s recommended in the ACMG guidelines. So they’re exactly the same in that case. And their recommendation is 50 base pairs from the penultimate exon junction and so that’s the cutoff we use as well.
Q: Is it possible to reuse previous assessments?
A: Yes, it most definitely is. That’s one of our main considerations, and one of the main advantages of VSClinical. Any assessments that you save in your report, you generated all that information gets saved in an assessment catalog. And if you come across the same variant again, or you come across a variant in the same region, those assessments will be used to inform the classification of that new variant.
Q: Do these workflows support CNVs?
A: Yes. The AMP workflow most definitely supports CNVs. In general, the oncogenicity scoring system doesn’t actually apply to CNVs, the idea here is that if you have a large CNV, then you’re going to most definitely want to walk this biomarker workflow and look for clinical evidence associated with that. But yes, the workflow most definitely supports interpreting and reporting on copy number variants as well as gene fusions and even wild considerations for wild type genes.
Q: Can I override the classification given by VSClinical?
A: You definitely can – at any time, any of these variants. If we classify it as likely oncogenic, but you’re not certain, you can always put that in your variant as uncertain significance. Or if you’ve got a variant of uncertain significance that you have reason to believe is benign and you disagree with our classification, you can choose to exclude that from the report entirely. Ultimately, we like to keep control completely in the hands of the clinician.
We hope you enjoyed this webcast recap. If you had a separate question not covered here, please give us a shout at info@goldenhelix.com and we will be more than happy to answer this for you. Thanks to everyone who attended, or tuned in for our recorded version, we look forward to seeing you on our next webcast!