One of the main goals of clinical genomic labs is to identify problematic variants in affected individuals. One tool to assist in this search is the phenotype driven variant ontological re-ranking tool in VarSeq called PhoRank.
A common situation facing clinicians is sorting through thousands of variants provided by an individual’s exome data (or possibly the individual’s nuclear family exome data) to try and determine which variant could be responsible for the individual’s illness symptoms in the shortest amount of time. Fortunately, this time constraint can be addressed with algorithms that incorporate phenotypic associations to highlight the most relevant genes with potentially damaging variants.
The PhoRank algorithm in VarSeq is based on some of the ideas of the Phevor algorithm published by Mark Yandell’s group in 2014. PhoRank and the Phevor algorithm use the phenotype search terms input by the user to determine which genes serve as highly relevant locations to start the variant investigation. Next, the shortest path between these starting locations and each of the sequenced variants for the individual in question is determined, and a gene score is assigned. This gene score also incorporates the gene and disease associations contained in different biomedical ontologies such as Human Phenotype Ontology (HPO) and the Gene Ontology (GO). A high gene score denotes a high relationship to the specified phenotype.
The variant based PhoRank algorithm can be applied in VarSeq by first annotating your variants with a gene annotation source (such as RefSeq Genes which comes included with VarSeq) and then selecting Variant PhoRank Gene Ranking in the algorithm selection menu.
To use PhoRank:
- Import your variants
- Annotate with a gene annotation source
- Select Add > Computed Data… (to see the dialog shown below)
- Select Variant PhoRank Gene Ranking under Gene > Project/Cohort
- Input phenotype(s) list
- *Note: Recent updates to the PhoRank algorithm now allow users to directly use the HPO IDs in conjunction with phenotype names, where before only phenotype names were accepted. For example, you can type HP:0000717 instead of “Autism”

The output to running the variant PhoRank algorithm provides the gene score, the gene rank compared to other genes, and the shortest path between the gene and one of the input phenotypes.
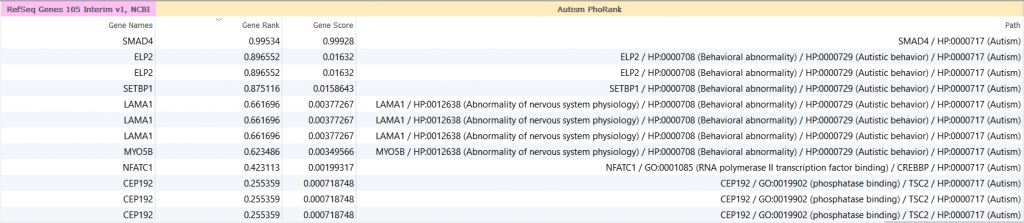
An example output is shown below using “Autism” as the phenotype for this analysis. The highest gene rank of 0.99534 belongs to the SMAD4 gene which is directly linked to the autism phenotype. The next entry in the list is the ELP2 gene which has a gene rank of less than 0.9 and multiple steps in the path. The decrease in gene rank value in this output denotes an increase in path steps between relevant genes in conjunction with a decrease in the phenotype-gene correlation relationship.
VarSeq variant PhoRank can be used to narrow the field of available search variants by ranking the relevance between a certain variant and specific phenotype and is especially useful for the single exome and family trio analyses that clinical diagnosticians commonly face.
For any additional questions about variant PhoRank Gene ranking or VarSeq, please reach out to us and we would be happy to set up a one-on-one demonstration.