VSClinical is a feature to evaluate clinically relevant variants according to the ACMG or AMP guidelines. This feature can also be used to identify if a variant has been observed previously or evaluate a manually inserted variant. Take, for example, the scenario where a colleague is interested to see if you have seen any variants associated with Bechet syndrome, which is a disorder that causes blood vessel inflammation throughout the body. Specifically, your colleague asks you if you have seen this variant: NM_006254.4(PRKCD):c.1352+1G>A.
Now searching for this variant might be difficult as you would need to have a strong recollection or go browsing through previous projects. That is unless you have VSWarehouse, which serves as a repository that can identify previously evaluated samples that contain the variant, what project it is present in, and at what frequency the variant is observed. However, if you don’t have VSWarehouse, you could instead use VSClinical to manually add that variant and search for hits using your assessment catalog.
To manually add a variant, you can open an existing project and go into VSClinical of a random sample. In the upper right-hand corner of the VSClinical evaluation, there is a Variants for (current sample) wizard (Figure 1). Clicking this option will open the dialog that shows all variants that have been imported for this sample, as well as the option to Add Variants.
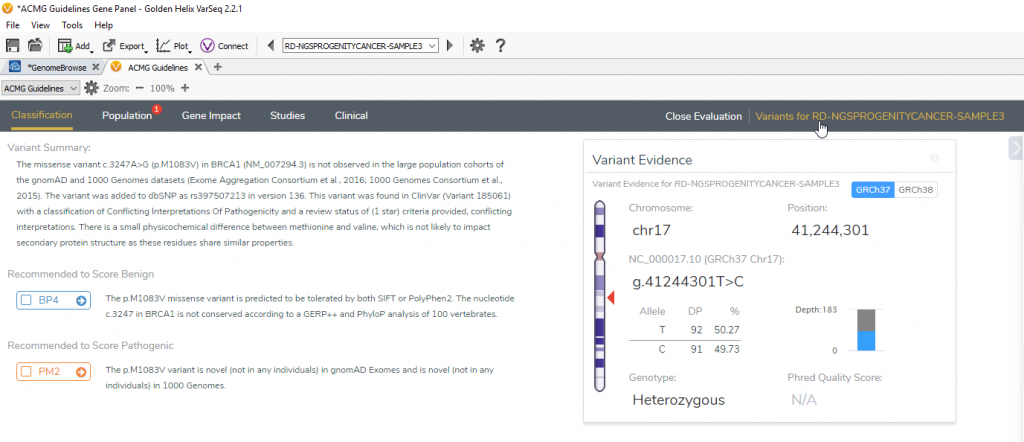
Figure 1: Variants for Current Sample option in VSClinical
If you select Add Variants, you will then be presented with a dialog where you can enter the variant of interest (Figure 2). In this case, I have entered NM_006254.4(PRKCD):c.1352+1G>A, which the colleague is interested in, but this can also support other nomenclatures. Once you select for the variant to be a heterozygous or a homozygous mutation, you can click Select. It will add the variant to the VSClinical interface after you select Add Variants. Now that the variant has been manually entered, we can then check for previous classifications as well as the associated details regarding the variant.
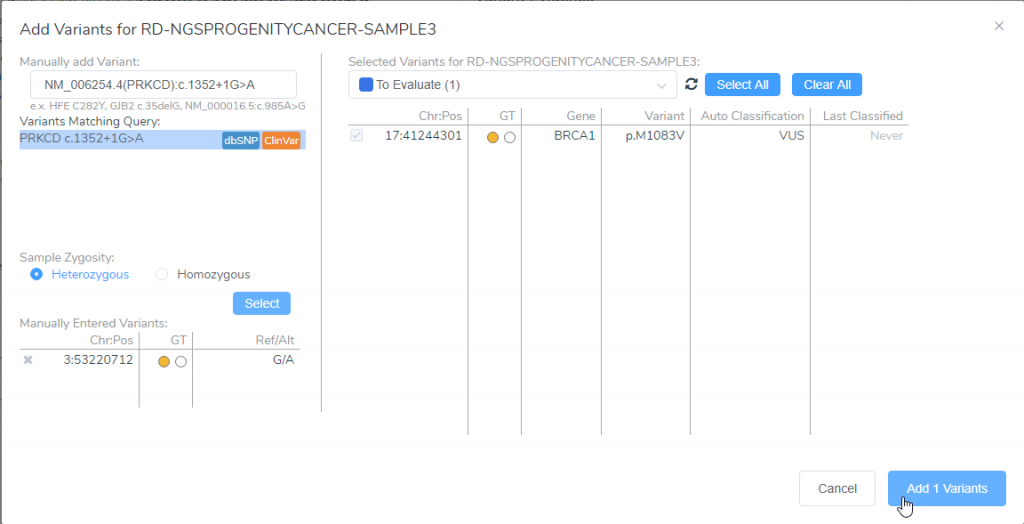
Figure 2: Manually adding variants to VSClinical
The first thing we can scroll to is the Interpretation section under the Classification tab (Figure 3). Here we can see that an interpretation has already been built for this variant, and it is associated with Autoimmune Lymphoproliferative Syndrome, Type iii. Furthermore, this variant was identified as Pathogenic, and it has been observed in two previous samples.
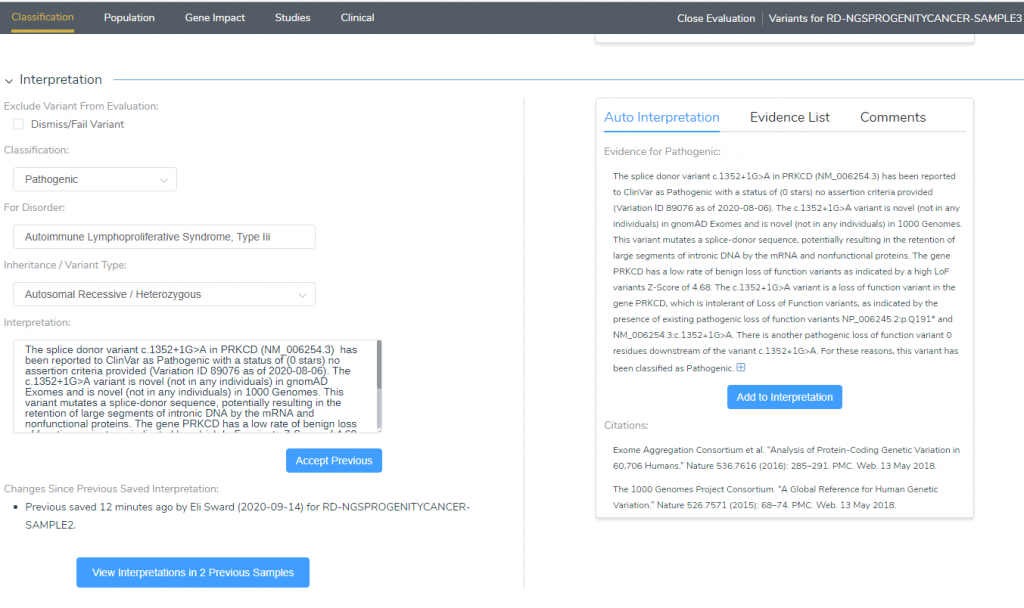
Figure 3: The Interpretation section will show the previous classifications of a manually added variant
If we wanted a more detailed explanation of how the classification was curated, we could then treat this variant as any other. For example, in the Population tab, we can look at the Tolerated Frequency section (Figure 4), which shows this condition as an autosomal recessive mutation. Furthermore, by clicking on the condition, we can see that it is associated with immune dysregulation. Although most phenotypes are variable, it is common to have significant lymphadenopathy associated with variable autoimmune manifestations.
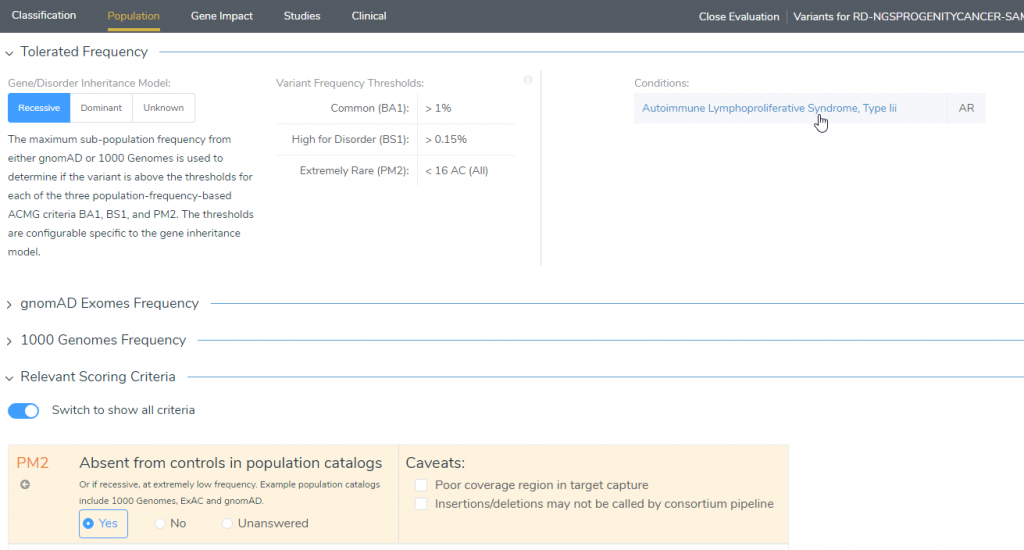
Figure 4: Clicking on the link for the Condition will provide a more in-depth description of the phenotype
If we then go into the Gene Impact tab under the Gene Region and Mutation Profile, you can look at neighboring variants present in ClinVar and your internal catalogs. Your internal catalog would be the assessment catalog created at the curation of VSClinical. In this example, there have been two previous samples that had this variant, which can be viewed by clicking on the icon (Figure 5). This will show you the date the sample was analyzed, the sample name, Genotype, Inheritance, Classification, Disorder, and Interpretation (Figure 6). This information can also be pulled up in the Studies tab in the Related or Nearby Variants section.
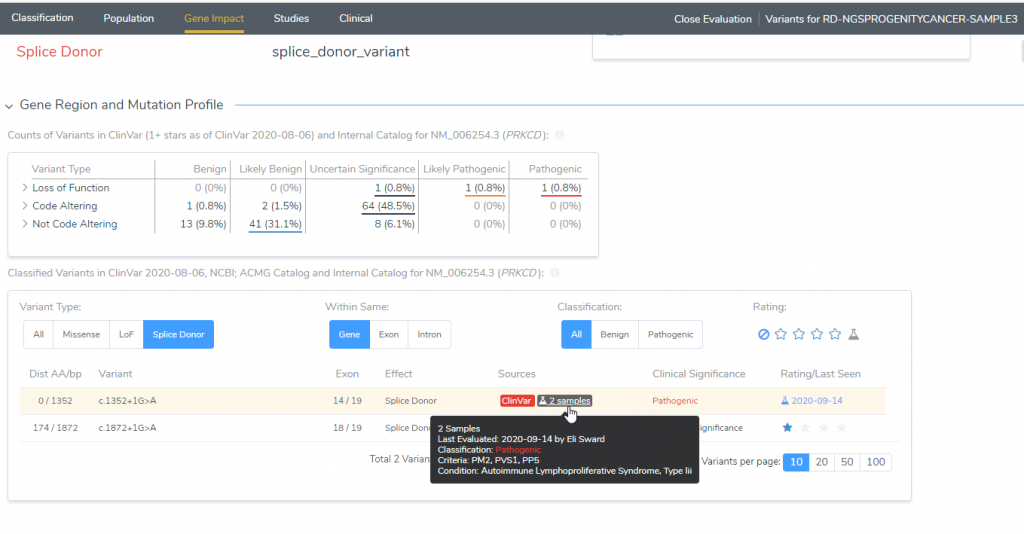
Figure 5: The Gene Region and Mutation Profile will indicate if the variant has been evaluated previously and what Clinical Significance was provided
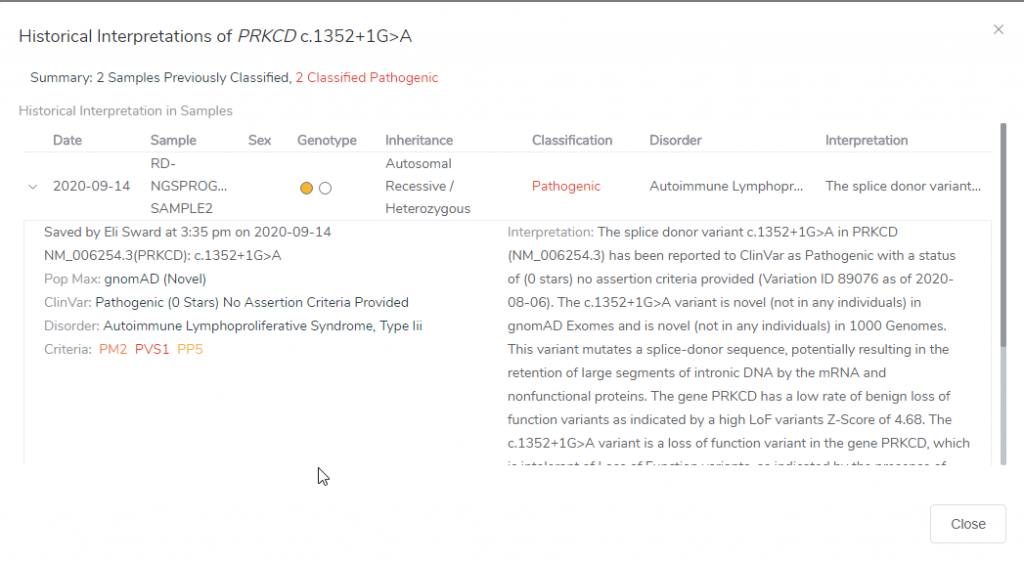
Figure 6: Historical interpretations for the variant of interest can be identified, and the sample it was observed in and the date
Lastly, users can also gain a deeper context of the variant by looking to see if there are ClinVar Assessments for the variant in the Studies tab (Figure 7). For this variant, there was a previous assessment, provided by OMIM, which was identified as a consanguineous marriage. This lead to the inheritance of the splice site mutation, leading to the patient experiencing recurrent infections and autoimmune disorders, including membranous glomerulonephritis and antiphospholipid syndrome. The patient’s father had Bechet Disease and mild autoimmune thyroiditis. This provides a great context of the variant as it shows previous classifications and summaries of the findings.
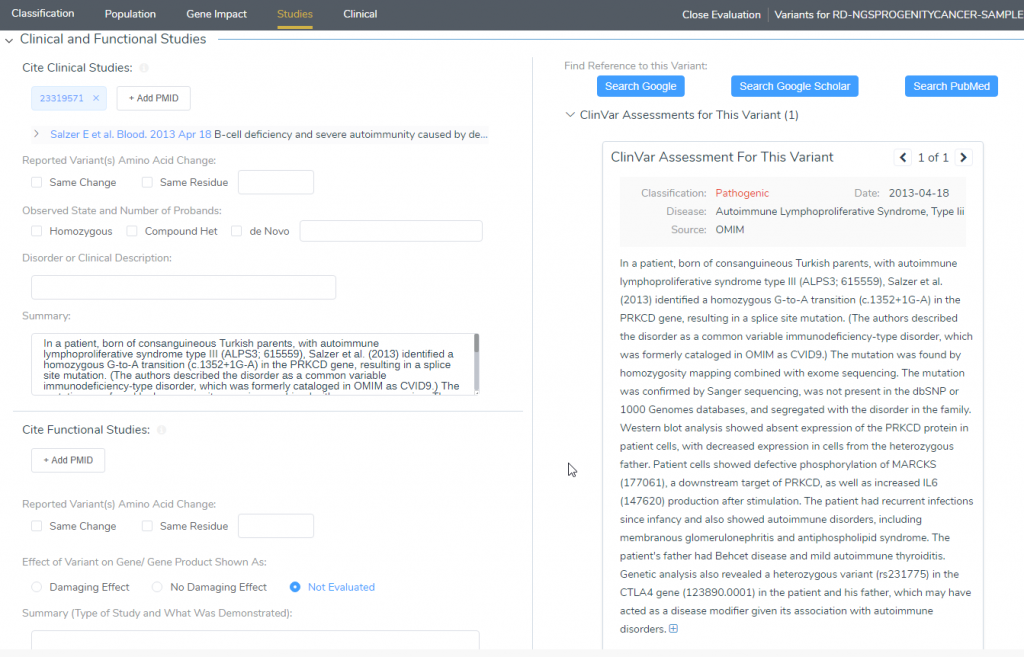
Figure 7: ClinVar Assessments identified that the variant resulted from a consanguineous marriage, and the father had Bechet syndrome
Using VSClinical as a variant search allowed us to see that the variant was observed in two previous samples. It was classified as an autoimmune disorder associated with the autoimmune lymphoproliferative syndrome and was classified as Pathogenic according to the ACMG guidelines. More importantly, this provided expert content to provide a conceptual framework regarding the variant and showed publications relevant to the variant. Coming back to the scenario, you would then be able to provide relevant information to your colleague, including how many samples you have observed the variant and what classification was created according to the ACMG guidelines.
Together, this is an example of how users can utilize current functionalities within VSClinical to identify if a variant has been observed in a previous sample. Using this feature might be a workaround to variant searches if you do not have VSWarehouse and is a great tool for determining the ACMG classification for variants not present in a VCF file. If you are interested in seeing VSClinical in action, you can email info@goldenhelix.com and one of our team members will be in touch!