As the number of genes on a gene panel increases, there is the possibility of picking up variants of medical significance that are not related to the primary indication for the test. Especially with large gene panels, exomes, and genomes, it is medically and ethically important to report variants that may be actionable to the patient. These include variants implicating hereditary risk of diseases as well as variants in genes associated with highly penetrant early-onset Mendelian disorders such as cardiomyopathy.
The ACMG Secondary Findings List (Incidental Findings)
Rather than every lab doing the research to determine which genes and classes of variants it should report as secondary findings, the American College of Medical Genetics and Genomics (ACMG) researched genes with associated disorders that met a number of criteria to be worth reporting to patients. This included evidence of actionability, severity, penetrance, and potential for therapeutic options or early screening recommendations to make a difference.
The first version of this list, sometimes called the ACMG56, was published in 2013 (updated in 2014) as a set of genes with “substantial clinical evidence that pathogenic variants result in a high likelihood of severe disease that is preventable if identified before symptoms occur.”
Since then, the list was updated in 2016 as ACMG SF v2.0 to an updated list of 59 genes. The process was improved for reviewing genes and based on updated clinical evidence and feedback one gene was removed and four added.
Very recently, the same incremental process was used to improve the gene list with the release of ACMG SF v3.0 as well as updated recommendations for how to use the list as part of a genetic test. The new list includes 73 genes and will continue to be updated by the ACMG Secondary Findings Working Group.
Integrating ACMG SF v3.0 Into a Variant Annotation and Filtering Workflow
It’s important to remember that not all variants detected in these gene lists should be reported. The original paper recommends “restricting the variants to be reported as incidental findings to those that meet criteria for reporting as Pathogenic”. Specifically, that includes variants previously established as a recognized cause of a disorder or by following the ACMG Guidelines for variant classification to establish the variant as the type that is expected to cause the disorder.
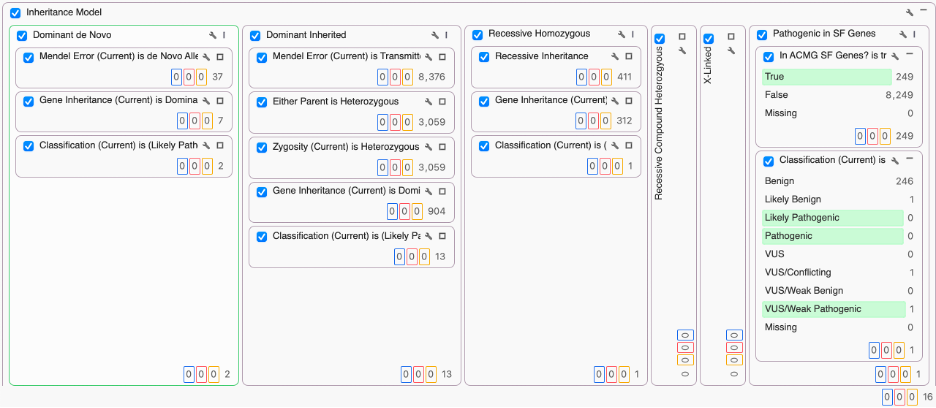
VarSeq has always included the ACMG SF list as part of the trio project templates. The “Match Gene List” algorithm supports providing a list of genes and annotating every variant on whether it is in one of the genes of the list. Combined with the ACMG Auto-Classifier algorithm to variants with evidence of Pathogenicity, it is possible to “fast-track” variants in a filtering workflow that should be evaluated as secondary findings alongside other workflow-specific filters.
With the release of VarSeq 2.2.4, we expanded our gene list filtering capability by adding support for managed gene panels. Gene panels can be used as part of built-in annotation algorithms, filters and as a component of the variant scoring and reporting process in VSClinical.
The ACMG SF v3 list is a built-in (system) managed gene panel in VarSeq v2.2.4. You can access it from Tools > Manage Gene Panels and open the dialog.
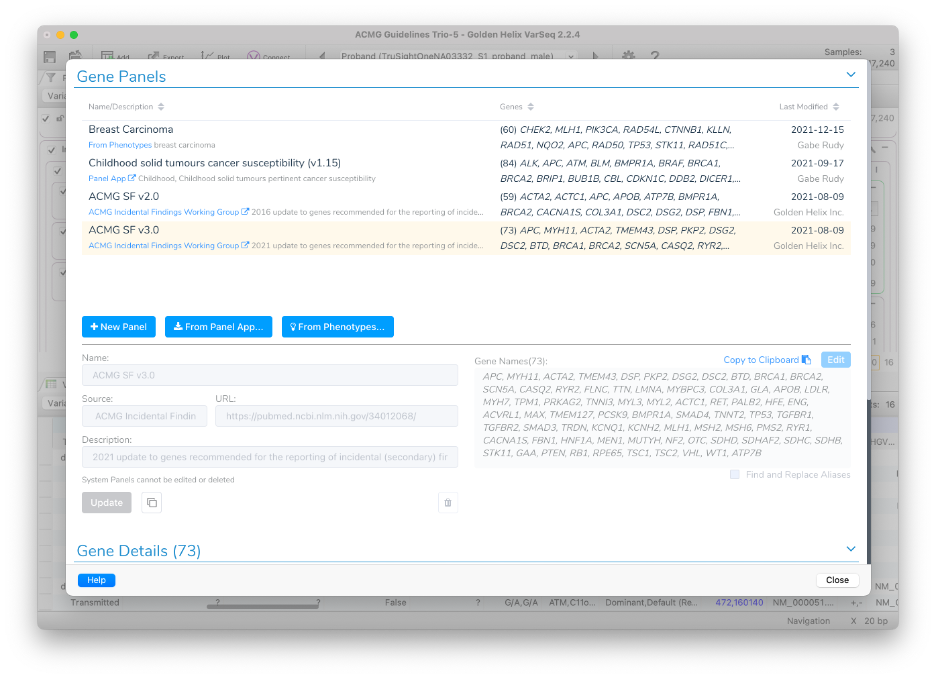
It is easy to leverage the built-in and custom gene panels from the VarSeq annotation and filtering context. There are several VarSeq features that allow selecting one or more gene panels managed by this dialog:
- Match Panels (Per Sample) Algorithm (Variants and CNVs): This algorithm uses a sample level field (provided by a manifest file on import) that lists one or more gene panels by name to be used for the current sample. For example, each sample may have associated ordered “tests”, each with a defined gene panel used to filter variants.
- Match Genes and Panels Algorithm (Variants and CNVs): Similarly, if the filtering is not sample specific, this algorithm creates a match column for variants that are in the genes of a selected panel. If the managed gene panel gets updated, newly created projects will use the latest list of genes.
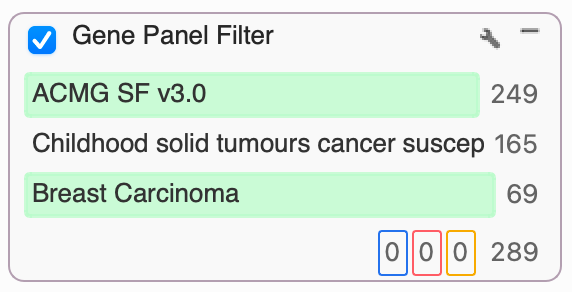
- Gene Panel Filter (In Filter Chain): This new card type allows configuring one or more panels to be displayed in the filter card and placed in any position in the filter chain.
- VSClinical: The VSClinical gene coverage tab can be configured to use a gene panel to define the genes that have coverage metrics computed, displayed and reported.
The field of medical genomics advances the understanding of the causative relationship between variants in specific genes and the implications to clinical care, and the ACMG Secondary Finings list represents the work of a lot of individuals to consolidate the knowledge of what a genetic testing lab can safely rely on as a baseline gene list for reporting incidental finding. With the advanced gene panel features in VarSeq v2.2.4, it is easier than ever to leverage this work.
To learn more about our products, VarSeq and VSClinical, please email us at info@goldenhelix.com.