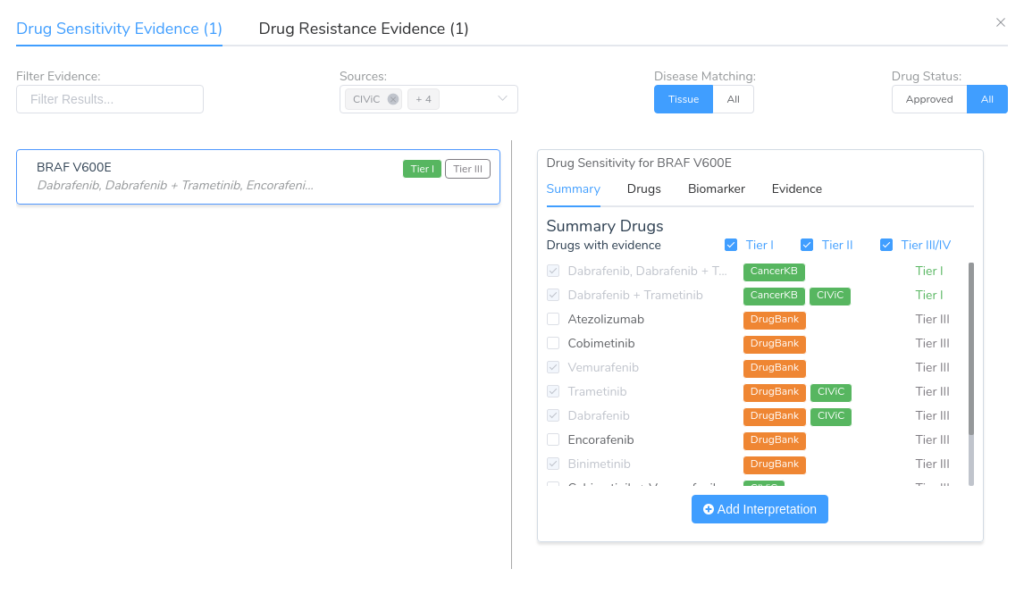
Discover the innovative clinical evidence search of the VSClinical AMP workflow, where patient biomarkers and tumor types are matched to the most relevant data from top sources like CIViC, DrugBank, and Clinical Trials.
One of the core features of the VSClinical AMP interpretation workflow is the ability to search third-party data sources to find clinical evidence that matches the patient’s biomarkers and tumor type. This includes evidence related to drug sensitivity and resistance, along with diagnostic and prognostic evidence.
To perform this search, VSClinical leverages a variety of third-party data sources including:
- CIViC: an open access resource for clinical interpretations of variants in cancer.
- DrugBank: a bioinformatics resource that combines detailed drug data with comprehensive drug target information.
- Clinical Trials: A curated database of clinical trials for cancer obtained from the NCI Cancer Clinical Trials and AACT databases.
Biomarkers
Performing queries on these data sources in VSClinical requires that each record be annotated with the relevant biomarker scope. To make this work smoothly, a lot of work is put in upfront to curate these data sources as annotation tracks that VSClinical and VarSeq access.
Curating raw data sources into structured biomarkers can be especially difficult for data sources like DrugBank, which do not provide specific information on known mutation associations. Even sources like CIViC, which are centered around cancer variant interpretation, have inconsistent variant naming conventions which make it difficult to determine the scope of a specific biomarker. For an illustration of this problem, simply look at the wide variety of ways various sources describe the variant BRAF V600E:
- BRAF p.V600E
- NM_001354609.1:c.1799T>A
- 7:140753336 A/T
- NC_000007.14:g.140753336A>T
We address this problem using a system of hand-written rules and patterns in conjunction with our powerful variant parsing engine. For a given term describing a biomarker, our curation scripts determine the mutation type by matching it against a series of over 50 different regular expressions. Once we have identified the type of biomarker, we can use our transcript annotation and variant parsing engine to identify the relevant region and extract the Ref/Alt for point mutations. Using this method, we can correctly identify a biomarker regardless of how it is described in a given source.
Tumor Types
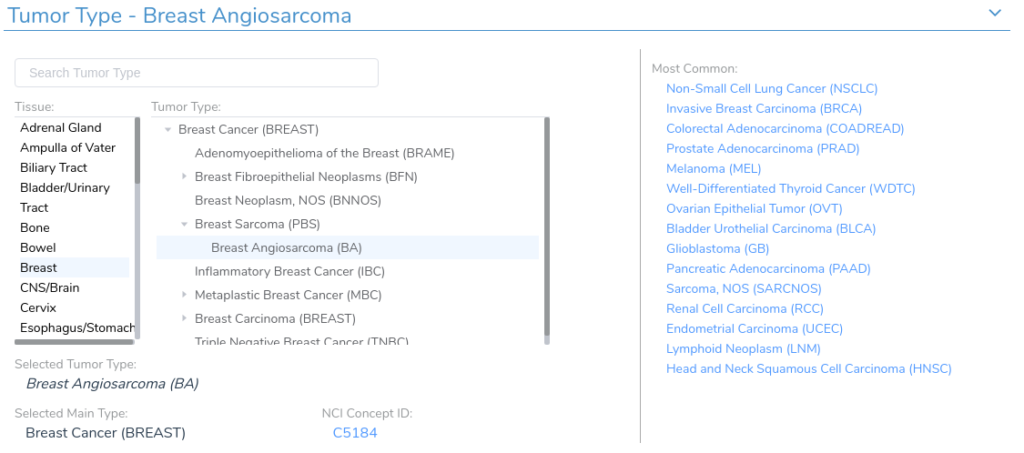
Another difficulty in the curation process is determining the appropriate tissue and tumor type for a given record. Our clinical evidence search relies on the VarSeq Cancer Ontology, which is a tree-structured cancer classification system in which root nodes represent general tissue types and branches represent different tumor type sub-classifications. Each VSClinical evaluation has an associated tumor type, which is used to identify relevant clinical evidence.
In order for our clinical evidence search to match the patient’s tumor type, each record in the queried sources must map to a specific tumor or tissue type in the Cancer Ontology. As with biomarker identification, identifying the correct tumor type can be challenging, as there are often many different ways to describe the same disorder. To address this, we have developed a search procedure to query the Cancer Ontology and identify the most relevant tumor type for a given disease description. Using this functionality, we can identify the correct tumor type for most records across the various evidence sources.
Conclusion
This is just a brief overview of the methods that we use to curate the various sources supporting VSClinical’s clinical evidence search capabilities. While data sources like CIViC and DrugBank are invaluable resources, significant effort is required to identify the biomarker and tumor type associations present in the data. Our curation team continues to make improvements to our curation process to ensure that our users have access to a comprehensive collection of clinical evidence data. For any questions regarding the VSClinical AMP workflow, please contact us at support@goldenadmin.