The world has been making a shift to use GRCh38 human genome reference coordinates, but the transition has not been fast. Many of the mainstay human catalog projects are changing to use native GRCh38 catalogs, or are remapping their current data to GRCh38 coordinates. While this seems to be the advancing goal, it is leaving researchers and analysts with the… Read more »
As many of our users know, VarSeq comes shipped with various project templates that are designed to give users a baseline workflow to get started with their projects. These templates are tailored for various applications including tumor-normal, trios, cancer and hereditary gene panels, and ACMG Guidelines workflows. The templates contain application-specific annotation sources and algorithms that will automatically load into… Read more »
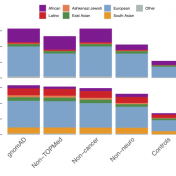
The Broad Institute team led by Dan MacArthur announced the release of gnomAD version 2.1 at last year’s ASHG conference. This new version boasted data from 125,748 exomes and 15,708 genomes, but the greater updates were the improved QC refinement and more discrete sub-population break downs. Although the majority of samples were counted in the previous 2.0.2 release, the additional… Read more »