VarSeq serves as a streamlined approach to handle the rare variant analysis typically carried out for your next-generation sequencing data. Our team at Golden Helix seeks to simplify this process by automating the curation of staple databases needed for filtering and evaluation of clinically relevant variants. The goal we seek to accomplish with our software is to provide ease in… Read more »
When it comes to clinical variant analysis for germline variants using ACMG guidelines, we understand that the clinical report is essentially the receipt for your services for the patient. In our recent webcast, Advanced Report Customization in VSClinical, we displayed several examples of report templates to show off the range of possibilities our users have to format their clinical report…. Read more »
If you stay current on the developments of Golden Helix features, you are aware of the substantial evolution of our copy number detection and evaluation capabilities in VarSeq. The process of CNV detection and evaluation is typically handled through the VarSeq graphic user interface. However, in some cases, users benefit from running this process via the command-line interface. Fortunately, Golden… Read more »
VCF file format comes with a lot of interesting quality assurance and statistics fields that can be used for filtering in VarSeq. Open your files in a text editor to see all the fields that are available in your files, each field will have a header line with a description of its content. See the VCF Specifications to help with… Read more »
We at Golden Helix love seeing our software and solutions out in action. Whether it is to find a clinical solution or to assist in academic research, seeing YOU use our tools to the best of their ability gets us excited to continue developing industry-leading software. For the 2022 Innovation Awards we would like to see all the creative and… Read more »
We are very excited to announce that just last week, we released VarSeq 2.2.4! In the past few months, we have been building the excitement for the 2.2.4 version of VarSeq with several webcasts in which we describe some of the headlining features in detail such as the new support for Gene Panels and Gene Lists, PhoRank Clinical, and Customized… Read more »
In February 2020, the American College of Medical Genetics (ACMG) and the Clinical Genome Resource (ClinGen) published a joint consensus on standards for the interpretation and reporting of copy number variants (CNVs) ranging from large CNVs spanning multiple genes to small intragenic events1. The guidelines consist of over 80 different criteria which are arranged into five distinct sections. These extensive… Read more »
Thank you to everyone who attended Using Golden Helix CancerKB to Accelerate NGS Cancer Testing! I had a great time showing off Golden Helix CancerKB and how it can enhance NGS analysis in cancer, I certainly hope you enjoyed it! If you were unable to make the live session, we have added the on-demand recording to our site, or we… Read more »
As a newer member of the Golden Helix FAS Team I, like many of our customers, went through a phase of learning the ins, outs, and shortcuts of clicking around the program. In an effort to expedite new users coming up to speed, or to teach some tricks to experienced users, I’ve compiled a list of the top things I… Read more »
The VarSeq CNV calling algorithm, VS-CNV, is a powerful tool for calling CNVs from the NGS coverage data stored in your BAM files. However, before this algorithm can be deployed in a clinical setting, it must be tuned and validated using data that is representative of your lab’s NGS workflow. In the past, this validation process could be difficult, as… Read more »
As we move towards the end of the year, our FAS Team is excited to announce our short blog series highlighting some of the Customization Features in our up-and-coming VarSeq release! The goal of this blog series is to show examples of how generating a clinical report can be customized to accommodate a wide range of functionality. Our December webcast,… Read more »
The customer-published articles this October cite the range of Golden Helix’s VarSeq annotation range and capability. The following publications feature everything from annotating an Italian nobleman mummy, assisting in identifying mutations in primary congenital and juvenile glaucoma, new mutations associated with muscular dystrophinopathy, and germline variants associated with head and neck cancer. In each of these cases, VarSeq was utilized… Read more »
Recently we have released blog posts discussing updates to annotations in VarSeq such as ClinVar and COSMIC. Keeping with that trend, in this blog post, I will discuss the most recent updates to the Golden Helix CancerKB database. For those how may be unfamiliar with the Golden Helix CancerKB source, it is a professional curated set of interpretations for the… Read more »
In addition to Golden Helix providing easy-to-use genomic software, we also provide value to our users by automating the curation of public databases used in our tools. These annotation sources serve multiple purposes not only leveraging key fields in the filter chain but also automatically supplying evidence for the ACMG and AMP classifications reviewed in VSClinical. Supporting the curation of… Read more »
In order to thoroughly assess a variant’s pathogenicity, it is important to take into account the variant’s effect on splicing. While the interpretation of variants that disrupt the pairs of bases at the beginning of a splice site is fairly straightforward, variants resulting in the introduction of a novel splice site are more difficult to interpret. In this blog post,… Read more »
With the release of VarSeq 2.2.4 just around the corner, I want to detail some new 2.2.4 features that will enhance somatic variant annotation and fusion analysis within VSClinical. A webcast back in July showed some of these updates in action, so if you are looking for some more content on this topic, I would highly recommend checking out the… Read more »
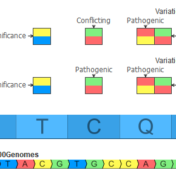
In the September 2021 monthly update to our curated ClinVar track, we made some changes that will result in roughly another 7,000 Likely Pathogenic and Pathogenic variants being available for annotation and use in the ACMG auto-classification system. Consensus Between Labs ClinVar has nearly one million unique variant classification records that are curated into multiple annotation tracks used in VarSeq and VSClinical on a monthly basis. Clinical… Read more »
Established in 2004 and headquartered in Chennai, India, with regional centers across the country, LifeCell runs India’s largest stem cell bank and has also diversified into diagnostics and tissue therapeutics. They employ roughly 2,000 people, providing genetic services to customers in the mother and baby space. Phani Nagaraja Setty is working as a scientist at LifeCell. Setty obtained his Master’s… Read more »
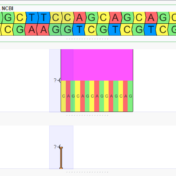
Merging variant records, VCFs, across samples is important when performing trio or family analysis as it ensures that hereditary relationships can be properly inferred. There are many ways to represent a single variant. Insertions and deletions may be right or left aligned, prefixes and suffixes can be added, and adjacent variants in the same sample may be combined or split… Read more »
Welcome to the August edition of our customer publications blog post! Each month we spotlight a few recently published articles by our incredible Golden Helix customers. With users spanning both research and clinical spaces, the topics vary widely across many fields. This month, we will be highlighting VSClinical users and the guided workflow. Host Genetics and Antiviral Immune Responses in… Read more »