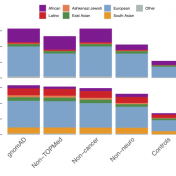
The Broad Institute’s release of gnomAD v4 needs no introduction as the data in this release is highly sought after by professionals in the genetics community, and the v4 release has a lot to boast about! The v4 release is roughly five times larger than the v2 and v3 releases combined and includes data from 807,162 total individuals. Naturally, exome… Read more »
We are excited to announce the release of our gnomAD v4 annotation tracks for VarSeq. This version of the GnomAD database represents a significant leap forward, including data from an impressive cohort of over 800,000 individuals — a remarkable 5x expansion compared to the previous releases. Notably, this dataset is comprised of two distinct callsets: exome sequencing data from 730,947… Read more »
In this blog post, I am very excited to talk about The Broad Institute’s release of the latest version of gnomAD, v 3.1.2, which is now available for use as an annotation source in your SVS or VarSeq projects. For VarSeq users, I also want to point out that gnomAD v3.1.2 can also be used as a population frequency in… Read more »
The Genome Aggregation Database (gnomAD) is a resource developed by an international coalition of investigators, with the goal of aggregating and harmonizing both exome and genome sequencing data from a wide variety of large-scale sequencing projects (1). We have covered this annotation in-depth in other blog posts, but this resource contains over 125,000 exome sequences and around 16,000 whole genome… Read more »
The Broad Institute team led by Dan MacArthur announced the release of gnomAD version 2.1 at last year’s ASHG conference. This new version boasted data from 125,748 exomes and 15,708 genomes, but the greater updates were the improved QC refinement and more discrete sub-population break downs. Although the majority of samples were counted in the previous 2.0.2 release, the additional… Read more »