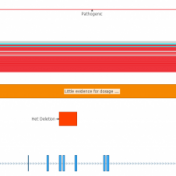
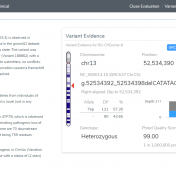
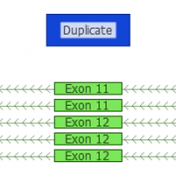
In a recent webcast, users were exposed to some new features upcoming in the next release of VarSeq. In this update, we will use an example de Novo CNV in a cardiomyopathy panel in VarSeq. There is a long list of new tools and polishes to the software but one major upgrade is the inclusion of the ACMG and ClinGen… Read more »
In this blog update, I’ll be walking you through some of the advanced plotting capabilities with GenomeBrowse. The strategy with any next-gen sequencing analysis is to filter down to interesting variants for either research or clinical conclusion. Golden Helix produces powerful software specifically tailored for this efficient and comprehensive search for interesting and clinically relevant variants. One additional advantage of… Read more »
In our recent webcast announcing the upcoming release of VarSeq VSClinical and the implementation of the ACMG guidelines for NGS CNVs, we had a number of live questions we didn’t get a chance to cover at the end of the presentation. I will follow up on those questions in this blog post. But first, if you didn’t get a chance to join us for… Read more »
It doesn’t take much effort to find articles discussing the value of Next-Generation Sequencing (NGS). There is a consistent tone amongst authors that implementing NGS pipelines are critical for clinical efficiency in both hereditary disorders and somatic. However, NGS strategies do not come without their own challenges. Challenges include not only the detection and calling of high quality/probability variants from… Read more »
Copy Number Variation (CNV) is a type of structural variation in which sections of the genome are duplicated or deleted. Although CNV events are rare in the human population, constituting approximately 10% of the human genome, they are also associated with being causal mutations for disease phenotypes. Because of this, it is important for clinical and research settings to identify… Read more »
The common approaches to detecting copy number variants (CNVs) are chromosomal microarray and MLPA. However, both options increase analysis time, per sample costs, and are limited to the size of CNV events that can be detected. VarSeq’s CNV caller, on the other hand, allows users to detect CNVs from the coverage profile stored in the BAM file, which allows you… Read more »
We love when our viewers send questions in during the webcast but unfortunately we can’t answer all of them during the time allotted! If you asked a question see below for answers, or if after viewing, you have any questions that weren’t asked, please feel free to send those over to support@goldenhelix.com. Does this work for FFPE derived DNA or ctDNA?… Read more »
With this two-part blog series, users should now be able to perform CNV analysis using their data, set up basic quality filter standards to isolate high-quality events and utilize annotations to hone in on publicly known events as well as in-house recorded CNVs from previous projects.
2017 was a busy year regarding the development of our CNV tools. Since the release of the CNV caller, we have produced quite a bit of content tailored to assist our users with getting started. Here are some links: Robarts Research Institute CNV analysis on patients with familial hypercholesterolemia CNV annotations Common CNV questions CNV calling with shallow whole genome… Read more »
One of our main focuses in 2017 was VS-CNV which allows clinicians to directly call CNVs in target regions quicker, easier and more affordably than CMA or MLPA testing. Our clients at Robarts Research Institute shared their recent publication with me which confirms that our time and dedication to our CNV capabilities was well worth it. I am delighted to… Read more »
September 27, 2017 12:00 PM, EDT While Copy Number Variants are important to detect and interpret in many clinical genetic tests, labs have been without a comprehensive solution that integrates the annotating and reporting of high-quality CNV alongside their existing NGS variants. Golden Helix has developed and validated with our clinical partners a specialized NGS-based CNV caller capable of detecting… Read more »
An Example of an Integrated Clinical Workflow for CNVs and SNVs In this blog series, I discuss the architecture of a state of the art secondary pipeline that is able to detect single nucleotide variations (SNVs) and copy number variations (CNVs) in one test leveraging next-gen sequencing. In Part I, we reviewed genetic variation in humans and looked at the key… Read more »
Examples of CNV Calling What do CNV calls actually look like? What are some of the key metrics to determine an event? Part IV of the Secondary Analysis 2.0 blog series will answer these questions by walking through some examples of how our CNV caller, VS-CNV, identifies CNVs. Golden Helix integrates multiple metrics to determine if a CNV event is… Read more »
Detection of CNVs in NGS Data Our Secondary Analysis 2.0 blog series continues with Part III: Detection of CNVs in NGS Data. We will give you an overview of some design principles of a CNV analytics framework for next-gen sequencing data. There are a number of different approaches to CNV detection. The published algorithms share common strategies to solve the… Read more »
In this blog series, I will discuss the architecture of a state of the art secondary pipeline that is able to detect single nucleotide variations (SNV) and copy number variations (CNV) in one test leveraging next-gen sequencing. In Part I, we reviewed genetic variation in humans in general and looked at the key components of a systems architecture supporting this… Read more »
Human genetic variation makes us unique. On average, humans are to 99.9% similar to each other. Understanding in detail what the nature of the difference in our genetic make-up is all about allows us to assess health risks, and eventually enables Precision Medicine as we determine treatment choices. Furthermore, it enables scientists to better understand ancient human migrations. It gives… Read more »
VarSeq will soon provide CNV exome analysis! In our webcast last week, we announced that we took our CNV caller, VS-CNV, to the next level. Along with the ability to call CNVs at the single-exon level in NGS gene panels, VarSeq can soon be used to call large loss of heterozygosity (LOH) and CNV events at the exome level. The combination… Read more »
ASHG 2016 is in our rear mirror. Again, it was bigger and better than the previous year. The conference hosted over 9,000 visitors from 66 countries. This gave the event a level of vibrancy that was evenly matched by the wonderful ambiance of the city of Vancouver. Nestled in between the two conference centers was a little pier offering spectacular… Read more »
Copy Number Variants have been important to clinical genetics for quite a while now. So, what has made now the right time to be looking at calling CNVs from NGS data? Well, there are a number of good reasons. The dominant one is simply that the NGS data you are already creating for calling variants can be used in many cases… Read more »