We are very excited to announce that just last week, we released VarSeq 2.2.4! In the past few months, we have been building the excitement for the 2.2.4 version of VarSeq with several webcasts in which we describe some of the headlining features in detail such as the new support for Gene Panels and Gene Lists, PhoRank Clinical, and Customized… Read more »
In February 2020, the American College of Medical Genetics (ACMG) and the Clinical Genome Resource (ClinGen) published a joint consensus on standards for the interpretation and reporting of copy number variants (CNVs) ranging from large CNVs spanning multiple genes to small intragenic events1. The guidelines consist of over 80 different criteria which are arranged into five distinct sections. These extensive… Read more »
In order to thoroughly assess a variant’s pathogenicity, it is important to take into account the variant’s effect on splicing. While the interpretation of variants that disrupt the pairs of bases at the beginning of a splice site is fairly straightforward, variants resulting in the introduction of a novel splice site are more difficult to interpret. In this blog post,… Read more »
Welcome to the August edition of our customer publications blog post! Each month we spotlight a few recently published articles by our incredible Golden Helix customers. With users spanning both research and clinical spaces, the topics vary widely across many fields. This month, we will be highlighting VSClinical users and the guided workflow. Host Genetics and Antiviral Immune Responses in… Read more »
Clinical diagnostic efforts in next-generation sequencing are commonly defined at a gene panel level. The validation process of adding new genes to any diagnostic panel is ongoing, but labs typically construct and validate their clinical workflows for the current status of verified genes. This is not limited to primary finding results but can also include any incidental findings among the… Read more »
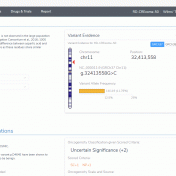
This blog post will cover an exciting new VSClinical feature in the upcoming VarSeq release. The ACMG Previously Interpreted Variants feature allows users to integrate databases of expert-curated variant interpretations into their VSClinical workflows. These data sources store variant-level interpretation data, including the classification, associated disorders, interpretation text, and scored criteria for each variant, along with notes providing a justification… Read more »
We are excited to share our latest publication with The Journal of Precision Medicine, “Implementing the ACMG Guidelines for CNV in a Commercial Software Solution”. “In 2020, ACMG in collaboration with the ClinGen working group developed a new set of guidelines for the clinical interpretation of CNVs. While theseguidelines provide a robust set of rules for interpreting intragenic deletions and… Read more »
The collaboration between the Clinical Genome Resource (ClinGen) consortium and the American College of Medical Genetics (ACMG) recently developed published guidelines for the interpretation of CNVs called on next-generation sequencing data. These new guidelines are the first to provide a robust set of rules for the interpretation of small intragenic deletions and duplications and are now automated in VSClinical. … Read more »
This morning I released a new version of my eBook “Clinical Variant Analysis – Second Edition.” The clinical interpretation of variants in Next-Gen Sequencing is a quickly evolving field. While the body of knowledge is growing exponentially, experts have to derive sound, clinical decisions leveraging an ever-expanding set of specialty databases, clinical publications, and algorithms that are designed to predict the… Read more »
In this blog post, I will be analyzing a loss-of-function splice variant in MTHFR using VarSeq. In the search for clinically relevant variants contributing to rare disorders, efficient filtering strategies are an important step in eliminating disinteresting variants. However, any applied filters must also ensure no interesting variants inadvertently get filtered out. Golden Helix provides the tools to complete this… Read more »
Those of you who have been attending our recent webcasts have learned about our upcoming VarSeq release. A part of that release will be an additional algorithm that will annotate variants matching the current sample. If you are not familiar with these webcasts, here are several on-demand webcasts I recommend to get you familiar with these new features: Evaluating Copy… Read more »
In the webcast, Evaluation of Copy Number Variants with VSClinical’s New ACMG Guideline Workflow, we discussed how VSClinical implements Section 4 of the ACMG guidelines. Specifically, we focused on integrating literature and publications to assess the pathogenicity of a CNV event when there was a lack of dosage sensitivity information. One of the primary pieces of evidence for evaluating genes… Read more »
Golden Helix is excited to release an upcoming VSClinical feature that allows users to analyze next-generation sequencing (NGS) CNV event reporting with ACMG guidelines. This feature will be the first in the NGS workspace to allow this capability and if you are curious about the functionalities you can get a sneak peek by looking at some of our most recent… Read more »
Our previous webcast from VP Gabe Rudy in September exposed us to some fundamentals of this years’ updated Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). This recent webcast was dedicated to breaking down these new guidelines… Read more »
A common discussion with our customers includes the challenges with the tertiary stage of analyzing next-gen sequencing data. This is the stage where all data from gene panels, exome, or whole genome scale pass through filters to quickly isolate the clinically relevant variant contributing to a patient disorder. Golden Helix has recognized these challenges in the scale of data and… Read more »
VSClinical is a feature to evaluate clinically relevant variants according to the ACMG or AMP guidelines. This feature can also be used to identify if a variant has been observed previously or evaluate a manually inserted variant. Take, for example, the scenario where a colleague is interested to see if you have seen any variants associated with Bechet syndrome, which… Read more »
In our recent webcast announcing the upcoming release of VarSeq VSClinical and the implementation of the ACMG guidelines for NGS CNVs, we had a number of live questions we didn’t get a chance to cover at the end of the presentation. I will follow up on those questions in this blog post. But first, if you didn’t get a chance to join us for… Read more »
The detection and interpretation of Copy Number Variants (CNVs) is vital for the clinical evaluation of individuals with a wide range of disorders. Golden Helix has remained at the forefront of CNVs in Next-Gen Sequencing (NGS) data since 2016 with the release of VS-CNV, our solution that allows you to both detect and analyze CNVs directly from NGS data. Earlier… Read more »
Golden Helix software provides huge analytic gain in handling large-scale genomic data. For example, a number of VarSeq users run cohort projects of whole genome level data processing hundreds of millions of variants at a time. However, many of our users are running gene panel level data for custom panels related to cancer (both hereditary and somatic), autism, cardiac, and… Read more »
It is common knowledge that variants can be germline or somatic depending on whether the variant was inherited or acquired after birth. A well-known example is cancer-causing mutations in the BRCA genes, wherein the mutation may or may not have been inherited. Understanding the origin of the cancer-causing mutation is important when assessing potential treatment options as well as identifying… Read more »