The last blog in this series covered streamlining variant analysis for large genetic cohorts, namely case-control studies, on a single-project basis. The reality when dealing with big data is that you often do not handle a high volume project all at once. Therefore, we will follow up on the topic of cohort analysis by discussing Golden Helix’s solution for streamlining variant analysis and tracking high-throughput projects over time and across batches of samples. Streamlining variant analysis for sample batches is mainly achieved through the use of our assessment catalogs and our genomic data repository VarSeq Warehouse.
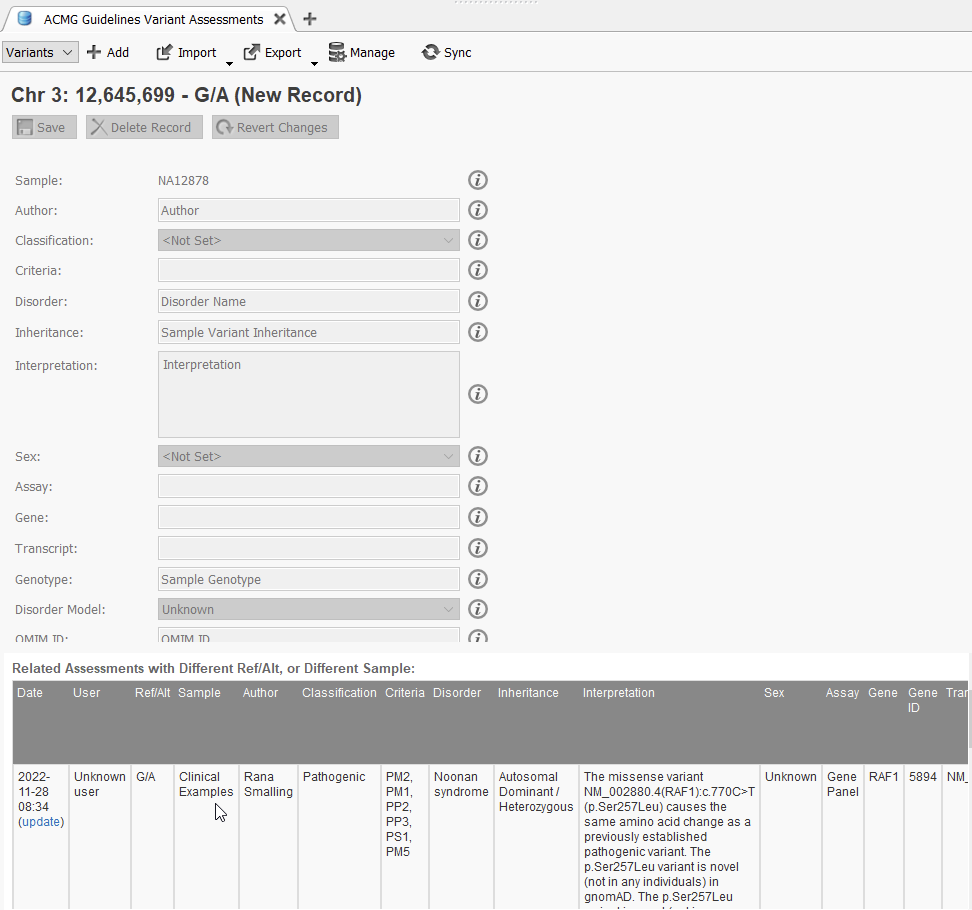
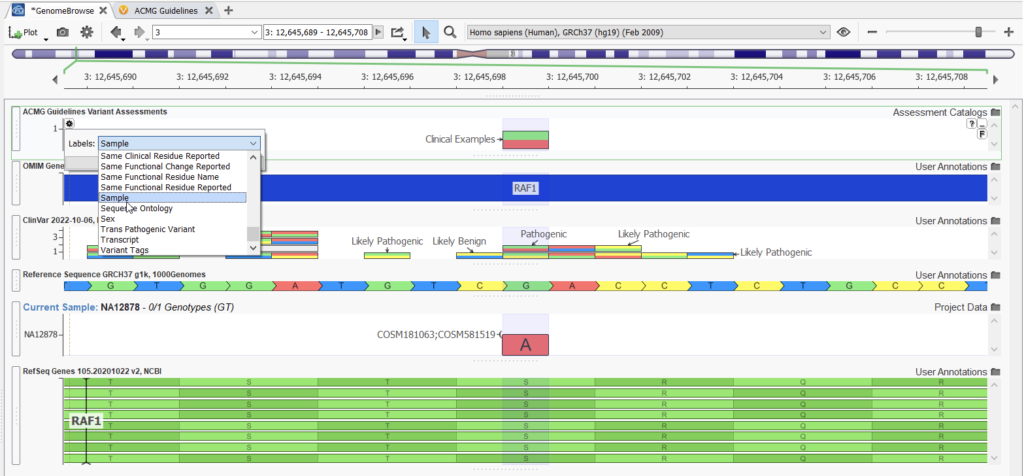
The VarSeq Assessment catalog feature allows you to save your variants into an assessment catalog for tracking at a later date or from another project or sample (Figure 1). In this example, the p.Ser257Leu variant in the gene RAF1 was encountered in sample NA12878, and based on our assessment catalog ACMG Guidelines Variant Assessments, we had seen this variant in another sample, Clinical Examples, and it was saved to the catalog by the user, Rana Smalling, after evaluation in VSClinical on 11-28-2022. This use case of assessment catalogs is limited to those stored in your local folder.
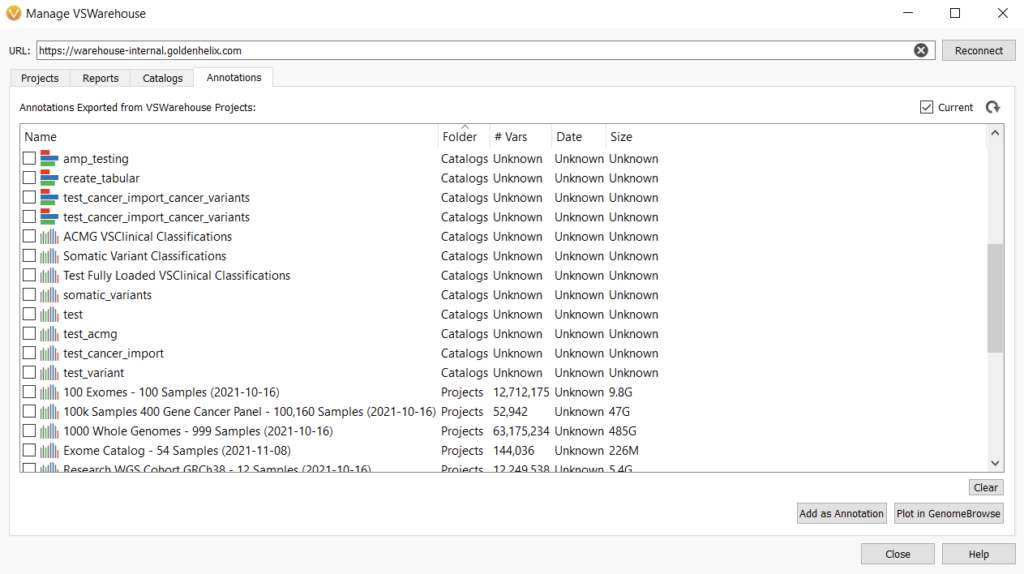
Once the variants in a project are saved to an assessment catalog, the catalog can be accessed again from the user’s locally stored assessment catalogs and plotted in Genome Browse. This will give a user an easy way to visualize variants that have already been encountered in another sample. In an assessment catalog, you can save information about the variant, such as a variant interpretation, classification, affection status, associated disorder, author, sample it was found in, and so on. You can use these fields to label the variants whenever you plot the assessment catalog (Figure 2).
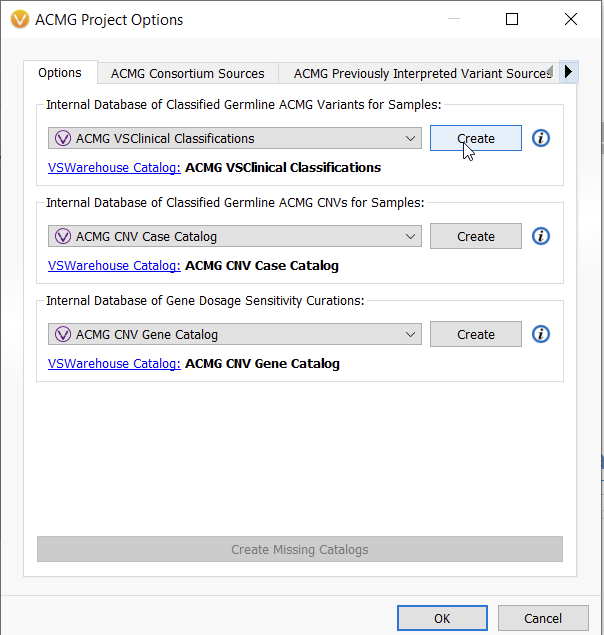
Assessment catalogs can be created for SNVs and CNvs, and more information can be found in the linked blogs. Our default assessment catalogs created for use with VSClinical are locally stored SQL-based catalogs, but users with the VarSeq Warehouse feature will be creating VarSeq Warehouse catalogs for storing variants in this genomic repository (Figure 3).
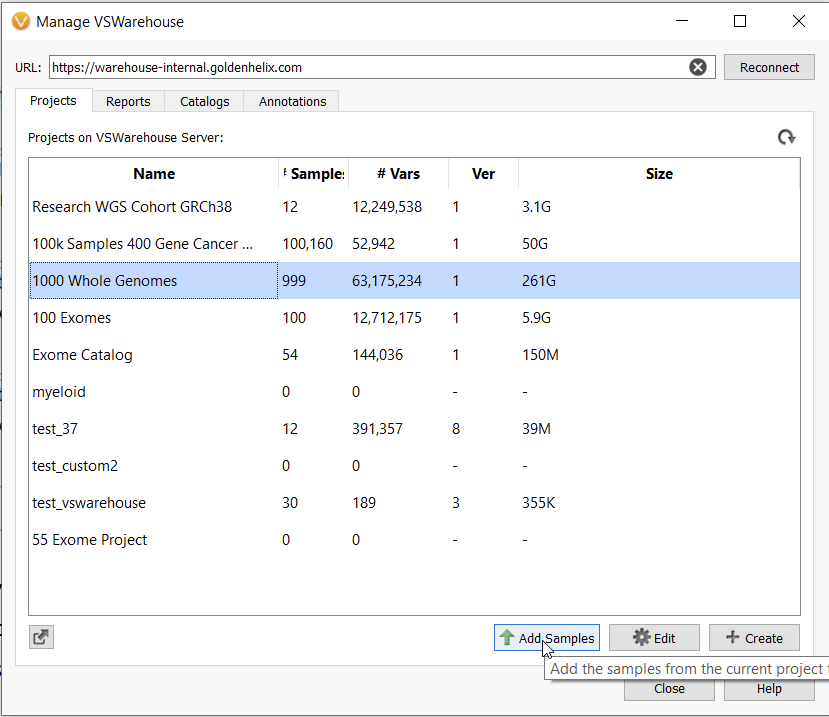
VarSeq Warehouse takes streamlining variant analysis for large genetic cohorts to another level. With Warehouse, users can access variant assessments and genomic data from any user on their team as long as they have been granted access. This genomic data can be used in a current project, for example, to plot and annotate with assessment catalogs stored in Warehouse by anyone on your team. Not only will you have access to assessment catalogs to use in this manner, but you can also use entire projects from Warehouse as annotations (Figure 4).
Think Warehouse is for you?
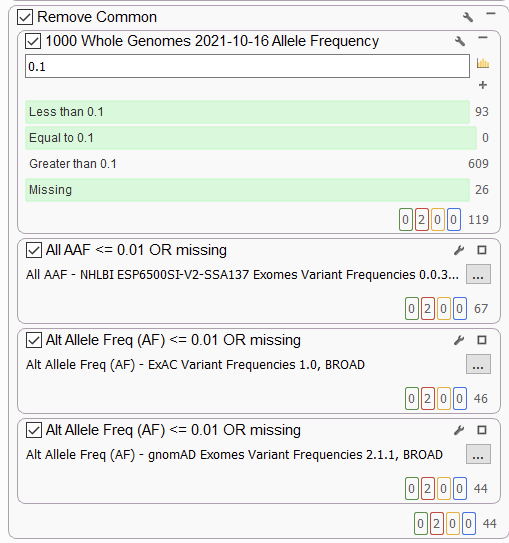
With this feature, you can leverage any information from variants in another project. Most importantly, you can leverage the allele frequency, as we saw in the last blog in this series, when using the Count alleles algorithm. This algorithm must be used whenever uploading a project to Warehouse so a user can keep track of allele frequency in a growing cohort of samples. From Warehouse, a user could pull fields like the alternate allele frequency in a large cohort project to use as a “Remove Common” variant filter (Figure 5). In this example, we used the 1000 Whole Genomes – 999 samples project seen in the list above.
Moreover, Warehouse can be used to add more samples into an existing project. This enables users to continuously grow a cohort and continuously calculate alternate allele frequency as they run and combine multiple batches of samples (Figure 6).
VarSeq Warehouse greatly simplifies the process streamlining variant analysis for large genetic cohorts. Warehouse can save the high-volume user much time and effort. Please feel free to reach out to support@goldenhelix.com for more information about assessment catalogs, VarSeq Warehouse, or if you have any questions about the contents of this article.
Golden Helix has developed innovative tools for simplifying variant analysis. Our suite, including Warehouse, assists in high-volume analysis. Being able to add more samples to an existing project enables users to grow an existing cohort and enables deeper analysis. Learn more by clicking the logo below.