Did you know you can control your preferred transcript settings for clinical interpretation in VSClinical?
Your lab is analyzing the DNA of a tissue sample from a patient with small cell lung cancer. The lab technician has imported the patient data into VSClinical to detect clinically relevant variants and evaluate and score these according to the AMP Guidelines, as well as find treatments or potential clinical trials for the patient. Fortunately, your lab has set up a workflow that keeps high-quality variants, searches genes, and regions that have submissions to the COSMIC database, and hones in on all variants that are not benign.
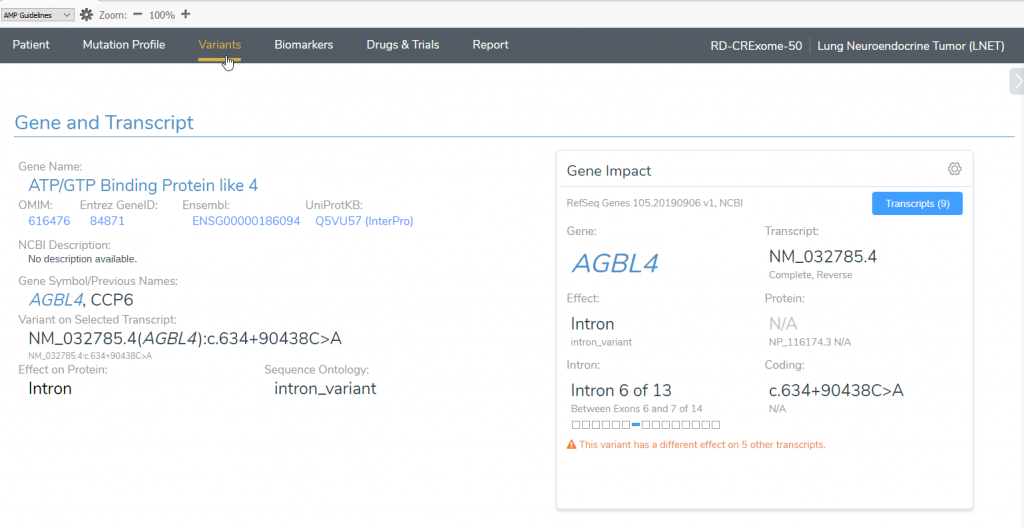
The lab technician finds a variant that is in a troublesome area wherein the variant of interest falls between two different genes with nine overlapping transcripts to choose from. In the image below, you can see that the heterozygous variant overlaps the BEND5 gene and in the intronic regions of the AGBL4 gene.
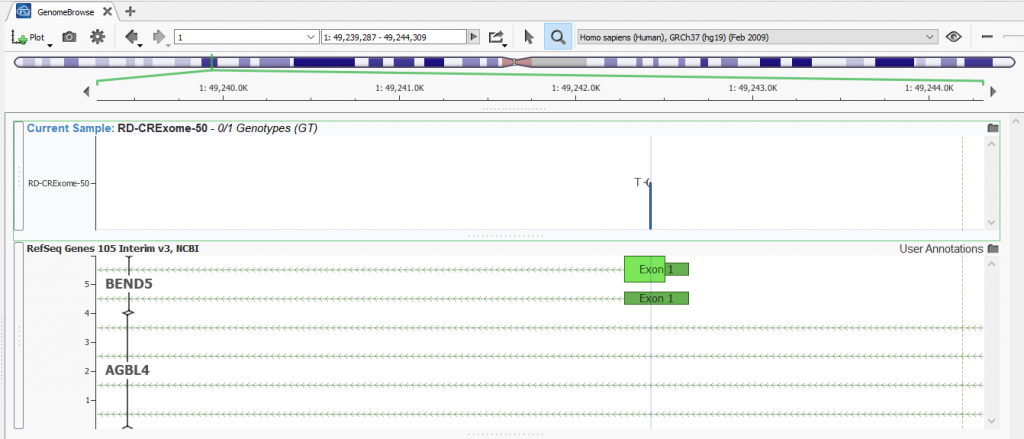
Figure 1: Heterozygous variant overlapping nine transcripts in the BEND5 and AGBL4 genes
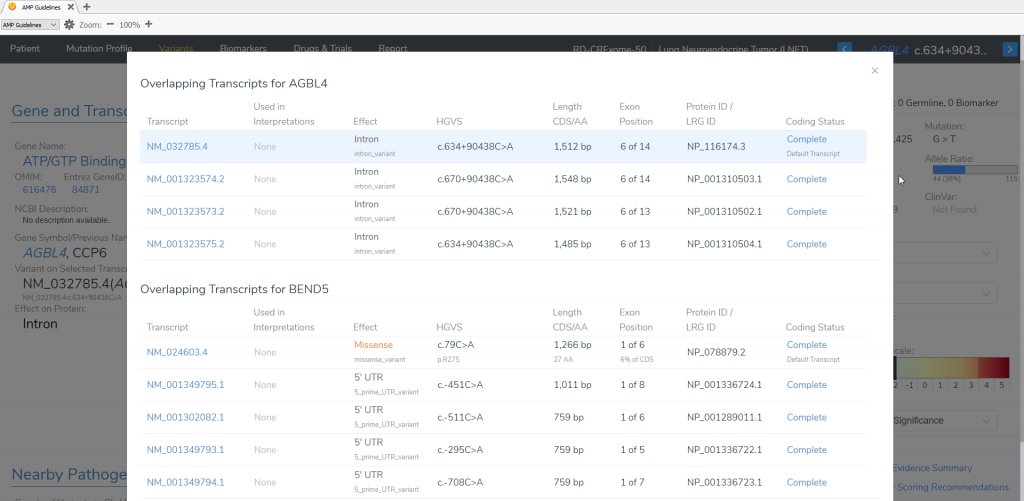
Fortunately, VSClinical captures and displays information for each of the transcripts, and within the AMP or ACMG guidelines workflows, you can select the transcript that is clinically relevant to the patient. After you include this variant to be evaluated according to the AMP guidelines, you see that the AGBL4 NM_032785.4 transcript is chosen as the default transcript. However, this transcript is not clinically relevant for the patient. In the image below, the VSClinical interface indicates that this variant has different effects on different transcripts (Figure 2).
Figure 2: AGBL4 NM_032785.4 transcript is displayed by default, but there is a notification in orange that the variant has different effects on other transcripts
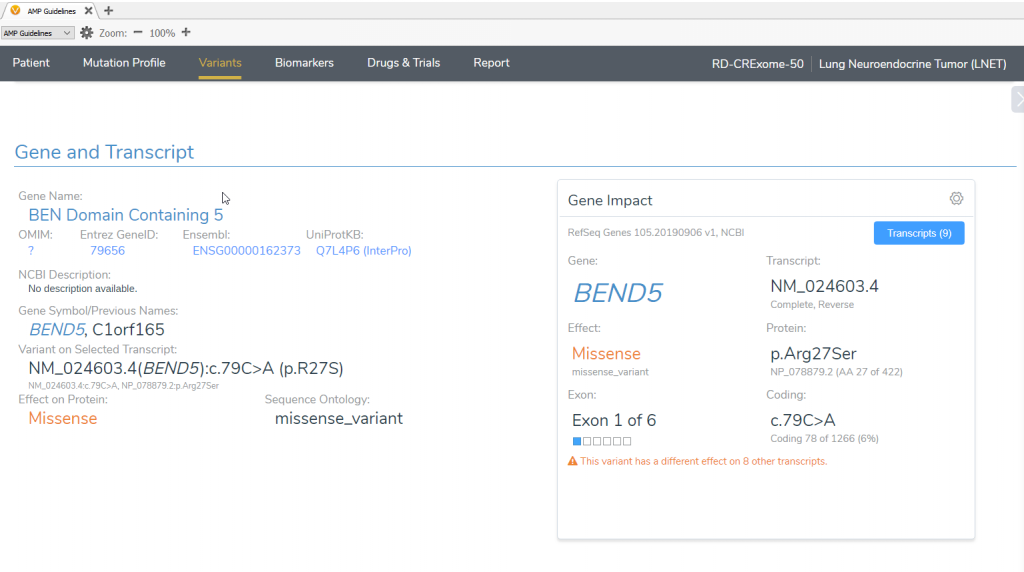
VSClinical captures and displays information for each of the transcripts and you can select the transcript that is clinically relevant to the patient, and small cell lung cancer, outside of what is considered the default transcript by RefSeq Genes (Figure 3). From the list of transcripts, you choose the transcript that applies to the patient, the BEND5 NM_024603.4 transcript.
Figure 3: VSClinical displays and allows you to select which transcript is clinically relevant for analysis
The VSClinical interface updates the variant interpretation according to the newly chosen transcript. Previously, the mutation had a synonymous effect and it was in the intronic regions of the AGBL4 gene. However, in the NM_024603.4 transcript of the BEND5 gene, the variant is a missense mutation that is potentially oncogenic and requires further investigation with the AMP guidelines (Figure 4).
Figure 4: The variant interpretation for the BEND5 transcript
Because the lab typically analyses lung cancer patient samples, it would be ideal if this transcript were chosen as the default transcript for analysis with future samples. VSClinical offers a few solutions for customizing your workflow, so you will not have to repeatedly select the transcripts that are clinically relevant for your lab!
There are two strategies you can use to set your own clinically relevant transcripts. The example described above eludes to one solution. Once you select a different transcript within the VSClinical interface as the new clinically relevant default transcript, VSClinical will save it as the new clinically relevant transcript for future evaluations if this variant turns up in other samples.
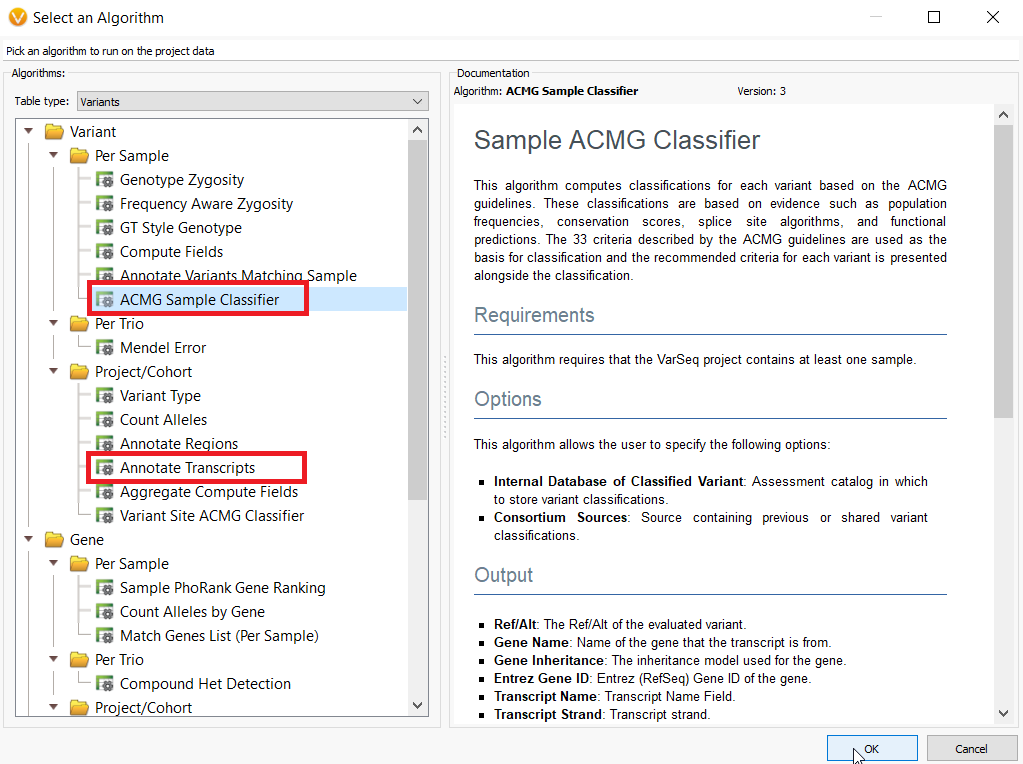
The second solution utilizes the Annotate Transcripts Algorithm or the ACMG Sample Classifier Algorithm within VarSeq to select clinically relevant transcripts in advance to be used in evaluation within VSClinical. These algorithms are particularly useful if you have a list of clinically relevant transcripts that the lab regularly analyzes that are different from default transcripts according to RefSeq Genes (Figure 5). To use this strategy, from within the VarSeq project interface, choose Add > Computed Data > Annotate Transcripts or ACMG Sample Classifier.
Figure 5: Annotate Transcripts and ACMG Classifier algorithms can be used to select clinically relevant variants for analysis with the ACMG or AMP Guidelines
Then, copy and paste the list of transcripts that you want to be considered as the default clinically relevant transcripts to be used in future evaluations within VSClinical (Figure 6). The dialog window in Figure 6 also shows that you can not only specify preferred transcripts, but also include splice site predictions, detect novel splice sites, and define splice site boundaries.
Figure 6: Dialog in which to enter the list of the desired clinically relevant transcripts and change splice site details
This blog was meant to describe a scenario wherein users may want to change the transcripts that are considered clinically relevant to the lab and then how users can incorporate those desired changes into VarSeq workflows. If you have questions or would like guidance on how to customize your pipelines further, our FAS team is always happy to schedule a training call and provide more hands-on assistance. Please reach out to support@goldenhelix.com if you would like a one-on-one training session or with any questions that you have!