Our team has returned from the annual meeting of the Association of Molecular Pathology (AMP 2019), and, as always, I am grateful for all the wonderful experiences we are bringing back with us. The plenary sessions and talks were bountiful, and we were very impressed with the well-organized exhibition. Hats off to everyone involved in planning this great event! Innovation… Read more »
We have recently added a tutorial to help introduce customers to the ease and utility of the AMP Guidelines incorporated in VarSeq’s VSClinical package. The AMP Guidelines allow users to sort through available clinical evidence in a streamlined fashion to arrive at final classification and interpretation and then transfer that information into a clinical report. And the AMP Guidelines also… Read more »
As I mentioned in the first part of this series, Sentieon allows users to call somatic variants against a matched normal sample and a tumor-only analysis. Utilization of a Tumor-Normal Workflow In addition to the fundamental process of alignment and variant calling, there are a few more steps that will improve the quality of your secondary analysis. Figure 1 (below)… Read more »
Golden Helix works to keep incorporating and updating great somatic annotation catalogs for our VSClinical users. We currently have the updated version of one of the largest cancer databases from the International Cancer Genome Consortium, or ICGC. Version 28 has been improved by integrating ClinVar and CIViC clinical annotations, and as always, increasing the number of mutations listed. The current… Read more »
The Golden Helix team is excited to be headed to Baltimore for the AMP 2019 Meeting, and we hope to see you there! You will find our team at booth #2856. Swing by or schedule a time to talk with us one on one about how Golden Helix’s platform is redefining next-gen sequencing analysis. Our team would love to hear… Read more »
VarSeq 2.2.0 was released today and this a stable release full of upgrades and polishes. Some of the newer features include the ability to store and include AMP Cancer assessment catalogs on VSWarehouse, quicker accessibility to common annotations plotted in GenomeBrowse, and the addition of all of our standard templates for the GRCh38 genome assembly. Many of the polishes were… Read more »
As our team returns from another successful ASHG conference, I would like to reflect on the great memories, connections, and future plans that were made at this meeting. First, I will start by thanking everyone involved with the superb planning and execution of the conference. We are thankful to have this opportunity to connect with our customers, partners, and friends… Read more »
The common approaches to detecting copy number variants (CNVs) are chromosomal microarray and MLPA. However, both options increase analysis time, per sample costs, and are limited to the size of CNV events that can be detected. VarSeq’s CNV caller, on the other hand, allows users to detect CNVs from the coverage profile stored in the BAM file, which allows you… Read more »
Before examining the clinical evidence associated with a specific mutation, a clinician must establish that the variant is likely to be a driver mutation which generates functional changes that enhance tumor cell proliferation. Our recent blog series “Following the AMP Guidelines with VSClinical” briefly mentioned how the oncogenicity scoring system in VSClinical could be used to automate and assist the… Read more »
From diagnosing, treating and preventing diseases in humans and animals, to preserving our global food supply, Golden Helix products are being used all over the world! In September, we were cited in many publications spanning a diverse range of species and topics. Below are brief recaps of a few of the studies where SNP & Variation Suite (SVS) and VarSeq… Read more »
Gene Fusion Background Gene fusions are hybrid genes that result from translocations, interstitial deletions, or chromosomal inversions that can lead to constitutive gene activation and result in increased or abnormal protein production. Increased or abnormal protein production subsequently can play an important role in tumorigenesis and thus identifying and evaluating this type of biomarker is important in the cancer workspace…. Read more »
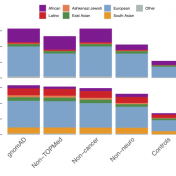
The Broad Institute team led by Dan MacArthur announced the release of gnomAD version 2.1 at last year’s ASHG conference. This new version boasted data from 125,748 exomes and 15,708 genomes, but the greater updates were the improved QC refinement and more discrete sub-population break downs. Although the majority of samples were counted in the previous 2.0.2 release, the additional… Read more »
We have covered a lot of ground in this Automating & Standardizing Your Workflows blog series. First, we saw how to perform secondary analysis with Sentieon to generate the necessary VCF and BAM files for tertiary analysis in Part I. The implementation of VSPipeline allowed for rapid import and project generation for a predefined cancer gene panel project template in… Read more »
VSClinical users can interpret and report genomic mutations in cancer following the AMP guidelines which we’re demonstrating in this “Following the AMP Guidelines with VSClinical” blog series. Part I introduced the hands-on analysis steps involved in creating a high-quality clinical report for targeted Next-Generation Sequencing (NGS) assays. We reviewed sample and variant quality, including the depth of coverage over the target regions by the sequencing performed for each sample. Now, we are ready to… Read more »
In our last part of this series, we showed how to run a pre-built workflow template via VSPipeline to automatically import and filter sample variants to streamline the search for clinically relevant variants. Now, we can deep-dive into our filtered, pathogenic variants to fully understand and capture their final classification and interpretation. Filtered Germline Variants for ACMG Guidelines The VSPipeline… Read more »
VSPipeline: Automating your Tertiary Workflows The first part of this “Automating & Standardizing your NGS Workflow” blog series covered the secondary analysis steps of read alignment and variant calling with Sentieon. The next step is to transition into the tertiary analysis via utilization of our workflow automation tool, VSPipeline. VSPipeline operates as a command-line tool meant to simplify the deployment… Read more »
In the world of genomics shaping precision medicine in oncology, the limiting factor is the time-to-sign-out of a fully interpreted molecular profile report. There are many components of the entire testing process that add to the turn-around time of each test. Many of these steps, such as sample prep, sequencing, and automated secondary analysis, are bounded and consistent in their time requirements. The hands-on… Read more »
Huntington’s Disease (HD) Background Huntington’s Disease (HD) is an autosomal dominant neurodegenerative disease that is caused by a mutation in the huntingtin (HTT) gene resulting in 36 or more CAG trinucleotide repeats in exon 1. Individuals affected by HD experience motor disorders including involuntary movements and poor coordination, cognitive impairments showing a decline in thinking and reasoning and psychiatric disorders… Read more »
Congenital Myasthenic Syndromes (CMS) History: Congenital Myasthenic Syndromes (CMS) are a group of rare hereditary conditions that can cause seizures, severe muscle weakness, respiratory problems, and potentially disabling weaknesses shortly after birth or early childhood (1). CMS is the result of abnormalities in acetylcholine proteins residing in the motor endplate of the neuromuscular junction (1). These abnormalities can be diagnosed… Read more »
Hypertrophic Cardiomyopathy History It was December 9th, 1989, when one of Loyola Marymount’s strongest inside players, Hank Gathers, collapsed during the middle of a collegiate level basketball game against UC Santa Barbara. Measuring in at 6’7” and weighing 210 pounds, Gathers was diagnosed with exercise-induced ventricular tachycardia, or in layman’s terms, an abnormal heartbeat. Even with the concerning nature of… Read more »