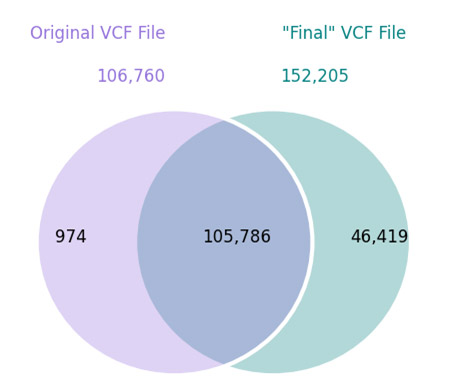
I’m a believer in the signal. Whole genomes and exomes have lots of signal. Man, is it cool to look at a pile-up and see a mutation as clear as day that you arrived at after filtering through hundreds of thousands or even millions of candidates. When these signals sit right in the genomic “sweet spot” of mappable regions with… Read more »
A few months ago, our CEO, Christophe Lambert, directed me toward an interesting commentary published in Nature Reviews Genetics by authors Bjarni J. Vilhjalmsson and Magnus Nordborg. Population structure is frequently cited as a major source of confounding in GWAS, but the authors of the article suggest that the problems often blamed on population structure actually result from the environment… Read more »
Last week Khanh-Nhat Tran-Viet, Manager/Research Analyst II at Duke University, presented the webcast: Insights: Identification of Candidate Variants using Exome Data in Ophthalmic Genetics. (That link has the recording if you are interested in viewing.) In it, Khanh-Nhat highlighted tools available in SVS that might be under used or were recently updated. These tools were used in his last three… Read more »
In 2011, I was looking for my next move and happened to attend a lecture given by Gabe Rudy at Montana State University. I was immediately struck by his passion and intelligence about the field of bioinformatics. He ended his talk by mentioning that Golden Helix was hiring, and I decided to apply. During my interviews, I discovered that these… Read more »
Presenter: Khanh-Nhat Tran-Viet, MHA, Manager/Research Analyst II at Duke University Date: March 7, 2013 Time: 12:00 pm EST, 60 Minutes Abstract Technological advances in next generation sequencing provide clinicians and researchers with more effective methods to identify pathogenic gene mutations for heritable diseases. To date, the National Eye Institute Bank lists over 450 genes associated with eye-related disorders. Analytical processing… Read more »
My investigation into my wife’s rare autoimmune disease I recently got invited to speak at the plenary session of AGBT about my experience in receiving and interpreting my Direct to Consumer (DTC) exomes. I’ve touched on this before in my post discussing my own exome and a caution for clinical labs setting up a GATK pipeline based on buggy variants… Read more »
In a recent GenomeWeb article by Tony Fong, “Sequenom’s CEO ‘Puzzled’ by Illumina’s Buy of Verinata, Lays out 2013 Goals at JP Morgan,” Harry Hixson, Sequenom’s CEO, expresses puzzlement over why its major supplier, Illumina, is acquiring a Sequenom competitor in Non-Invasive Prenatal Testing (NIPT), and thus apparently competing with one of its major customers. In a JP Morgan interview… Read more »
Twenty-four new variants discovered, each conferring more than a 2-fold risk of developing ASD Date: January 29, 2013, Noon EST, 90 minutes Presenters: Dr. Hakon Hakonarson, Director of the Center for Applied Genomics, Children’s Hospital of Philadelphia Dr. Mark F. Leppert, Professor of Human Genetics at the University of Utah and Chief Science Advisor at Lineagen Dr. Michael Paul, President… Read more »
We are excited to let you know about new scripts to aid in filtering rows/columns and identifying unique values in a column, as well as two updated scripts. Don’t forget about the Technical Support Bulletins which keep you up-to-date on all the latest script news. You can stream this feed via an RSS reader, receive email updates, or see the… Read more »
As a senior studying statistics at Montana State University, I was fortunate to be granted the opportunity to intern with the development team at a local software company. I was even more fortunate when this internship turned into full-time employment after I graduated. Working on a team with extremely talented individuals, I realized the significant impact our company makes through… Read more »
Happy new year! We hope all of our readers, clients, and users had a great holiday season filled with friends and family! While Golden Helix did take a break from blogging in December, we’ve still been pretty busy bringing you new and exciting things. We’re pleased to announce the addition of genome maps and annotation tracks for two new plant… Read more »
In preparation for a webcast I’ll be giving on Wednesday on my own exome, I’ve been spending more time with variant callers and the myriad of false-positives one has to wade through to get to interesting, or potentially significant, variants. So recently, I was happy to see a message in my inbox from the 23andMe exome team saying they had… Read more »
Join me on December 5th for a one-hour webcast as I explore my personal exome provided by the Exome Pilot project of 23andMe. Exome sequencing has seen many success stories in the realm of diagnosing highly penetrant monogenic disorders as well as in informing treatment of certain cancers. As the use of exome sequencing expands to more complex polygenic disorders… Read more »
After much anticipation, we are excited to announce that SVS 7.7 is available! It’s been a lot of work, but we are pleased with the result and know you will be too! SVS 7.7 includes: Improved RNA-Seq Analysis Package for mRNA Expression Profiling Import tabularized quantification data from the Golden Helix pipeline or other sources. Utilize DESeq to perform differential… Read more »
Golden Helix is proud to announce our participation in EA’s 2012 “Leave Your Fingerprint on the Cure.” This event will be taking place at ASHG 2012 in San Francisco at the Moscone Center on November 7th. The fifth annual “Leave Your Fingerprint on the Cure” was designed by EA to help raise money for researchers in their efforts to discover… Read more »
Recently, I have been spending some time analyzing real patient data. I’m preparing for a webcast I’ll be giving in which I will walk through the process of replicating the findings of Dr. Gholson Lyon‘s study on the novel disease diagnosis he named Ogden Syndrome. Being so close to data that comes directly from clinical settings got me thinking about… Read more »
Dr. Folefac Aminkeng is a Postdoctoral Fellow at The Centre for Molecular Medicine and Therapeutics (CMMT) at the University of British Columbia in Vancouver, BC, Canada. He utilizes GWAS studies to identify single-nucleotide polymorphisms (SNPs) that might be associated with serious adverse drug reactions (ADRs) in cancer therapeutics. The field of pharmacogenomics—how one’s genetic makeup affects drug response—has grown exponentially… Read more »
Date: Wednesday, October 24th, 12:00 pm EDT | 4:00 pm UTC Presenter: Gabe Rudy, VP of Product Development Abstract Next-generation sequencing of DNA has shown to be a successful tool to discover causal variants of rare, highly penetrant mendelian diseases. In this webinar, we will analyze clinical samples to demonstrate the bioinformatic workflows and visualization techniques used to reproduce the… Read more »
Since Dr. Ken Kaufman gave his webcast on Identifying Candidate Functional Polymorphisms in SVS, we’ve been working with Dr. Kaufman to simplify and automate many of the steps in his workflow. I touched on this in my last blog post, and I’m excited to report that with Ken’s help, we’ve been able to simplify the workflow even more. In particular… Read more »
Editor’s Note by Gabe Rudy: I’ve had the chance to exchange thoughts, emails, and blog post comments for a while now with Jeff as he has written posts on NGS Leaders and engaged with me on 23andMe. He has also worked with Golden Helix software as part of Dr. Todd Lencz’s research efforts at Zucker Hillside Hospital until he recently… Read more »