In our recent webcast, Advancing Agrigenomic Discoveries with Sequencing and GWAS Research, Greta Linse Peterson featured bovine data which she download from the NCBI website. The data was downloaded in SRA format and in order to analyze the data in SVS, the files had to be converted to BAMs and then merged into a single VCF file. Since many of… Read more »
At Golden Helix our number one priority is empowering genetic researchers world-wide with software tools that are as effective as they are robust. So needless to say, we are thrilled to announce a recent collaboration with the Ontario Genomics Institute (OGI), a not-for-profit organization focused on driving and catalyzing the life sciences industry in Ontario. Through this exciting partnership, we… Read more »
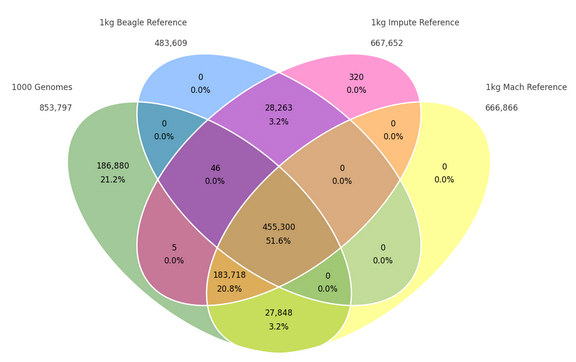
In a recent blog post (Comparing BEAGLE, IMPUTE2, and Minimac Imputation Methods for Accuracy, Computation Time, and Memory Usage), Autumn Laughbaum compared three imputation programs. Data can be exported from, or imported into, SVS in the standard file formats for these and other imputation programs. The goal of this blog post will be to review the different tools available to… Read more »
Tis the season of quiet, productive hours. I’ve been spending a lot of mine thinking about file formats. Actually, I’ve been spending mine implementing a new one, but more on that later. File formats are amazingly important in big data science. In genomics, it is hard not to be awed by how successful the BAM file format is. I thought… Read more »
Are you ready to show your research to the world? Do you and a colleague want a free one-year SNP & Variation Suite license? Could you use a new laptop? Then we have a contest for you! As part of our ongoing commitment to empowering genetic researchers around the world, Golden Helix is hosting a competition for abstracts. All academic, government,… Read more »
A report from the World Congress of Psychiatric Genetics Earlier this month, while much of the genetics community was scrambling to edit and print their posters for ASHG, I had the opportunity to attend WCPG, the World Congress of Psychiatric Genetics, in Boston. This was my second trip to WCPG and it is becoming one of my favorite events to… Read more »
Hey everyone! It’s time once again for the illustrious ASHG – this year in Boston, MA. We are very excited to get to see all of our colleagues and friends and hear about what you’ve been up to. This year we will have six in-booth (#618) demonstrations by Gabe Rudy, VP of Product Development, showcasing SVS 8.0: Identifying Candidate Functional… Read more »
Last week, we presented a webcast on Workflow Automation in SVS. If you were unable to attend, a recording of it is on our website: http://www.goldenhelix.com/Events/recordings/making-ngs-data-analysis-clinically-practical/index.html In this post I’ll respond to some of the questions we were unable to answer within the allotted time. Will you provide a link for the software used in the webcast? I used Golden… Read more »
Kellie Carey discussing her treatment with her doctor. Image by Jesse Neider for The Wall Street Journal Just a few weeks ago, the case of Kellie Carey made it to the front page of the Wall Street Journal. Initially, her prognosis in 2010 was very dire. Three months. Lung Cancer. As I write this article, Ms. Carey is still alive… Read more »
Presenter: Autumn Laughbaum, Biostatistician with introduction by Dr. Andreas Scherer, President & CEO Date: September 10, 2013 Duration: 60 Minutes Abstract Exploring next-generation sequence data requires an iterative process whereby a researcher can find a “needle in the haystack” that contributes to a particular disease or other phenotype. Once that needle has been found, a workflow can be established for… Read more »
Utilizing Identical Twins Discordant for Schizophrenia to Uncover de novo Mutations We are living in exciting times – the reality of high-resolution Cand individual genome sequencing now offers renewed hope in the search for the causes of complex diseases. When this technology is combined with genetic relationships, individual sequences add unrivaled proficiency. Our lab is located in London, Ontario, Canada… Read more »
Genotype imputation is a common and useful practice that allows GWAS researchers to analyze untyped SNPs without the cost of genotyping millions of additional SNPs. In the Services Department at Golden Helix, we often perform imputation on client data, and we have our own software preferences for a variety of reasons. However, other imputation software packages have their own advantages… Read more »
Presenter: Dr. Bryce Christensen, Statistical Geneticist Date: July 24, 2013 Duration: 60 Minutes Abstract Golden Helix GenomeBrowse™, a free visualization tool for all types of sequence data, was introduced in 2012 to broad acclaim. Researchers using GenomeBrowse discovered a product far beyond the status quo with seamless navigation of sequence alignments and other genomic data using a fluid, fast, and… Read more »
Humans are said to have a 0.1% rate of diversity, seemingly small, but actually quite significant when considering the future of personalized medicine. Gaining a deeper understanding of this genetic variance can help determine susceptibility to disease as well as medicinal response and outcomes. One-third of human genetic diversity can be found in a likely contender for the birthplace of… Read more »
In a couple of short weeks, Gabe is headed off to TCGC in San Francisco where he will be giving part of a short course. He was super excited about it last year and is even more so this year. I sat down with him yesterday to find out why. Jessica: What’s TCGC? Gabe: Last year I got to attend… Read more »
Thanks to everyone for the great webcast yesterday. We had over 850 people register for the event and actually broke the record! Take that Bryce and Gabe! If you would like to see the recording, view it at: Mixed Models: How to Effectively Account for Inbreeding and Population Structure in GWAS. While preparing for this webcast, we chose to focus… Read more »
Last month, Bryce Christensen wrote a little about mixed models and their application in GWAS. He promised that this analysis would be available in SNP & Variation Suite (SVS) “soon,” but didn’t elaborate. We are now excited to announce that three mixed model methods are available in SVS: GBLUP, EMMAX, and MLMM! To help demonstrate their utility and when to… Read more »
Presenter: Greta Linse Peterson, Senior Statistician Date: Wednesday, June 5th, 2013 Time: 12:00 pm EDT, 60 minutes Abstract Population structure and inbreeding can confound results from a standard genome-wide association test. Accounting for the random effect of relatedness can lead to lower false discovery rates and identify the causative markers without over-correcting and dampening the true signal. This presentation will… Read more »
Thirteen years ago, Dr. Robert Kleta had never heard of a genome-wide association study (GWAS), let alone considered doing one. Now, Dr. Kleta and his colleagues at the University College of London regularly publish articles in The New England Journal of Medicine and other journals on the genetics of rare diseases and their associated phenotypes. States Kleta, “For rare diseases,… Read more »
Time goes by fast. With the completion of the Human Genome Project in 2003, scientists worldwide were trying to understand the cause and effect of variations in the genome as they relate to functionalities, traits and disease. Along the way, we at Golden Helix helped researchers analyze data, discover variations and draw conclusions. It turns out that the real bottleneck… Read more »