In the new Genotype Imputation tool that is coming soon to SVS, allele encoding is an important part of matching data between the target and the reference panels. If the same platform provider is being used, then A/B encoding can be used. However, it’s better to use the Reference/Alternate allele encoding associated with AGCT format to ensure accuracy. If an… Read more »
Since we released our Phenotype Gene Ranking algorithm in VarSeq, it has become a staple of the way people conduct their analysis. It allows for a combination of filtering with ranking to prioritize follow-up interpretations of analysis results. Our PhoRank algorithm will be available in our upcoming SVS release to also aid in the numerous research workflows performed on SNPs… Read more »
Join our upcoming webcast – Clinical Reporting Made Easy Wednesday, February 15th 12:00 pm EST Clinical labs need to be able to process samples down to a short list of variants and publish a professional report. VSReports helps scientists and clinicians alike create timely, actionable reports that can improve clinical decision making and streamline patient care by seamlessly incorporating the… Read more »
It may have been easy to miss in the drum-beat of monthly annotation updates we do here at Golden Helix, but there are a couple of things that are very special about the January update to the ClinVar database: We added new fields including HGVS names of variants and citations in PubMed for variants ClinVar nearly doubled in size by… Read more »
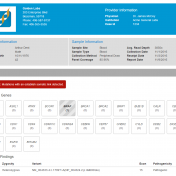
Clinical labs need to be able to process samples down to a short list of variants and publish a professional report. VSReports helps scientists and clinicians alike create timely, actionable reports that can improve clinical decision making and streamline patient care by seamlessly incorporating the results of tertiary analysis into a customizable clinical report. To include the VSReports functionality in… Read more »
Variant interpretation is an integral part of any workflow that results in some decisions being made about the validity and suspected functional impact of a variant in a given sample and their presenting phenotypes. The VarSeq Assessment Catalog functionality is designed to assist the VarSeq user in streamlining this process. To include this functionality in your workflow, you will first… Read more »
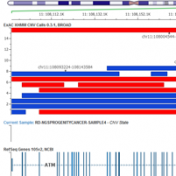
ExAC CNVs were released publicly with a recent publication, providing the full set of rare CNVs called on ~60K human exomes. While there are many public CNV databases out there, this is the first one that was derived from exome data, and thus includes both extremely rare and very small CNV events. With the recent release of Golden Helix’s CNV calling… Read more »
True to its nature, VarSeq offers multiple data export options. You can export result tables from VarSeq to Text, VCF, a VarSeq annotation file and most importantly an XLSX (Excel) File. VarSeq’s Excel export options provide a lot of flexibility in the information that is exported and preserve the formatting of data during the export process from VarSeq to Excel. This… Read more »
While clinical assessments of germline mutations have been collected in ClinVar under the stewardship of the NCBI and the collaborate effort of many testing labs, the same type of resource has been missing for mutations that could informal clinical care in Cancer. Or at least, that is what I thought until I started to work with CIViC. With the stewardship of… Read more »
This year we have added some very important features to our software including CNV calling in VarSeq and integrating our premium annotations into SVS. We have also made many improvements to our software’s performance so that you can handle growing datasets with relative ease. In an effort to thank our community for their support throughout the year, we are happy… Read more »
ASHG 2016 is in our rear mirror. Again, it was bigger and better than the previous year. The conference hosted over 9,000 visitors from 66 countries. This gave the event a level of vibrancy that was evenly matched by the wonderful ambiance of the city of Vancouver. Nestled in between the two conference centers was a little pier offering spectacular… Read more »
The new Annotate and Filter algorithm is now available with the release of SVS 8.6.0, see the release notes for full details on all new and updated features. To access this new functionality, you simply need to update your SVS installation to the new version. The update can be done by clicking the Update Available link at the bottom of… Read more »
In our SVS 8.6.0 release, we updated our Annotate and Filter Variants feature to utilize our powerful VarSeq annotations. Annotations can be run against gene, interval, variant, and tabular tracks, including RefSeq, ClinVar, CADD, OMIM and OncoMD. The new streamlined dialog allows users to select track specific options and to set up custom filters. While our public annotation repository has… Read more »
Gabe Rudy’s webcast yesterday, Big Data at Golden Helix: Scaling to Meet the Demand of Clinical and Research Genomics, was a huge success with well over 300 registered. In today’s blog post, I wanted to recap the Q&A session with Gabe. If you missed the webcast, check it out! Question: What is the end goal for an application like Warehouse? Answer: The… Read more »
Big data is here, but fear not, you don’t need a Hadoop cluster to analyze your genomes or your cohorts of tens of thousands of samples! It turns out, for the kind of algorithms employed in variant annotation and filtering, running optimized local programs is often faster anyway. As we support our diverse customer base, we have definitely seen the… Read more »
Question: Now that I’ve added annotation sources for my sample, filtered down to a list of interesting variants, flagged those variants and generated a clinical report, can I save or copy the annotation sources and filters for use on another sample? Short Answer: Yes! Long Answer: VarSeq was created with ease and efficiency in mind. In VarSeq, once you’ve defined… Read more »
Every month hundreds of clinicians and researchers access the variety of free resources on the Golden Helix website. Our resource library hosts eBooks, webcasts and tutorials to keep the community apprised of new methods, informed on best practices and to help our customers get the most out of their software purchase. Here is a list of the 5 most watched webcasts… Read more »
Since 1999, Bonei Olam has been providing large-scale funding for fertility treatment and research. The non-profit’s mission is to provide whatever means or resources necessary to help childless couples achieve the dream of parenthood. Today, it is recognized in the worldwide medical arena for its leadership role at the forefront of reproductive medicine, research and technology. Specifically, Bonei Olam has… Read more »
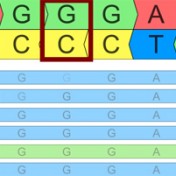
Variant Normalization: Underappreciated Critical Infrastructure It may surprise you to learn that every variant in the human genome has an infinite number of representations! Of course, although true, I’m being a bit hyperbolic to prove a point. Even seemingly simple mutations like single letter substitutions are legitimately represented differently in the local context of other mutations that can be described… Read more »
Whole exome sequencing workflows using SNP & Variation Suite (SVS) was presented in a recent webcast, by Dr. Robert Hamilton from the Hospital for Sick Kids. In particular, he performed some filtering on his data to look for only heterozygous variants in his sample of interest, removed variants with allele frequency less than 0.005% based off of the ExAC Variant Frequency… Read more »