This blog post will cover an exciting new VSClinical feature in the upcoming VarSeq release. The ACMG Previously Interpreted Variants feature allows users to integrate databases of expert-curated variant interpretations into their VSClinical workflows. These data sources store variant-level interpretation data, including the classification, associated disorders, interpretation text, and scored criteria for each variant, along with notes providing a justification for each applied criterion. If a variant under evaluation is included in one of these sources, the criteria, classification, and interpretation text will be automatically filled when the variant is added to an evaluation. The upcoming VarSeq release will include two expert-curated variant interpretation databases that have been preconfigured so that they can be added with the click of a button:
- ClinGen Expert Curated Interpretation of Variants
- IARC TP53 Database
In the following sections, we will cover both of these data sources in detail and show how they can be leveraged to streamline your interpretation workflow.
ClinGen Expert Curated Interpretation of Variants
The standards for the interpretation of sequence variants developed by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) have been embraced as best-practice guidelines for germline variant interpretation by clinical laboratories around the world. They have been vital for establishing a consistent methodology for interpreting germline variants. However, since the initial publication of the ACMG guidelines, there has been a need for modifications tailored toward specific individual genes and diseases. To address this, ClinGen has established multiple Variant Curation Expert Panels (VCEPs) to provide gene/disease level criteria specifications, evaluate the clinical validity of gene-disease relationships, and evaluate the pathogenicity of individual variants.
This track includes ACMG criteria recommendations, interpretations, and comments from several ClinGen Expert Panels covering diseases such as hearing loss, cancers, hereditary diseases, and more. The interpretations produced by these panels are included in the ClinGen Expert Curated Interpretations track, which is available in the VarSeq annotation source library. We are pleased to announce that in the upcoming VarSeq release, VSClinical will support the automatic classification of any variant included in this annotation source.
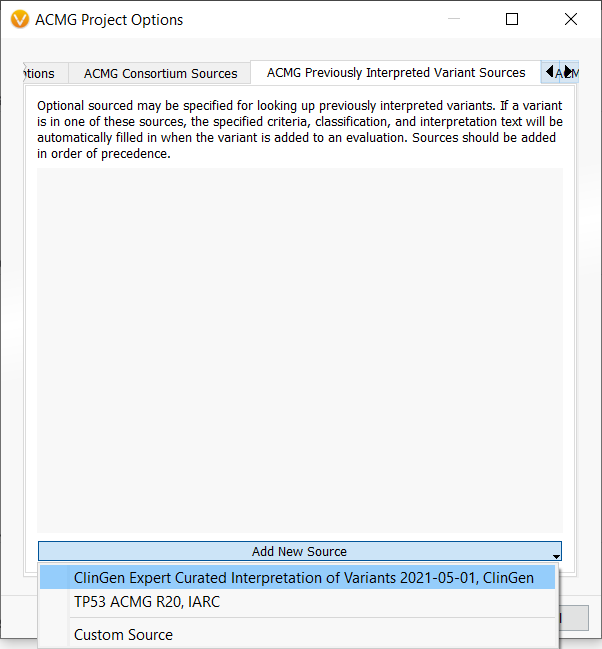
This functionality is enabled by VSClinical’s new Previously Interpreted Variant Sources feature, which can be accessed through the ACMG Project Options. After opening the ACMG Project Options by clicking on the gear icon in the ACMG tab, we navigate to the Previously Interpreted Variant Sources tab and use the dropdown to add the ClinGen Expert Curated Interpretations track.
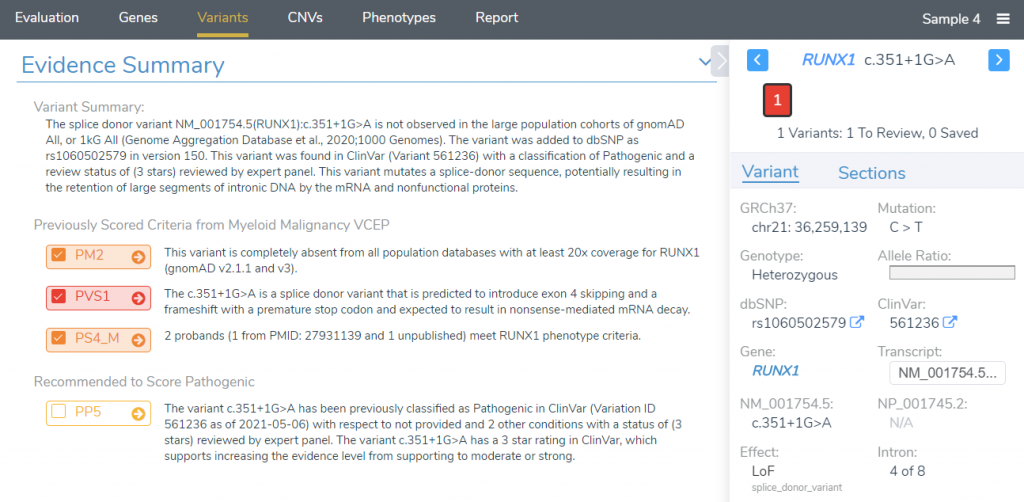
Once this track has been added, we are ready to begin interpreting our variant. For this example, we will be looking at the variant RUNX1 c.351+1G>A, a splice site mutation that disrupts the donor splice site of exon 4. If we look at the variant tab, we notice that several criteria have been automatically applied for the variant, and we already have a pathogenic classification. Our Evidence Summary section shows that these criteria have been applied from the ClinGen Myeloid Malignancy VCEP.
In the Evidence Summary section, we can see each scored criteria, along with corresponding comments taken from the ClinGen Expert Panel interpretation, providing the specific reasons why these criteria were scored. Notice that while PP5 was recommended by the VSClinical recommendation engine, it was not selected for this variant. If we click on the arrow next to PP5, we can investigate why these criteria were excluded. While there are no comments provided for this criterion, we can explore the recommendations of the ClinGen SVI working group by clicking See further discussions on PP5/BP6.
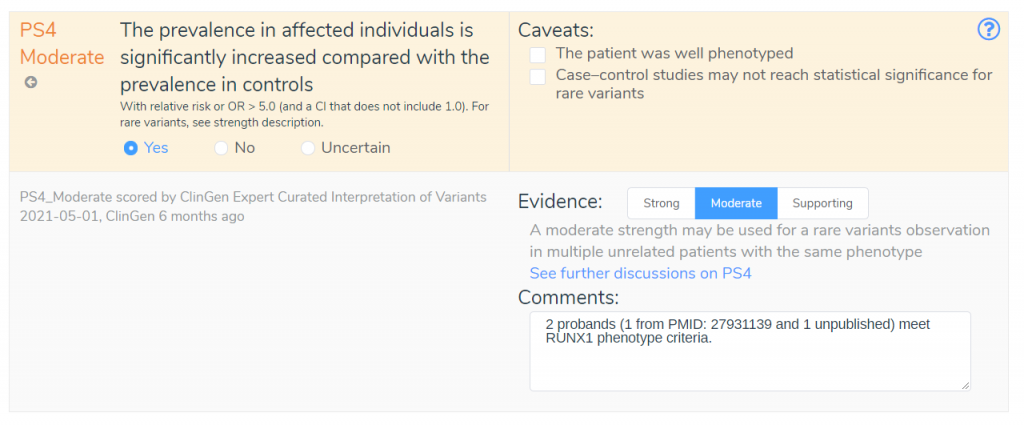
If we look at the discussion provided by the ClinGen SVI working group, we can see that they recommend criteria PP5 and BP6 be discontinued given the availability of primary data. Using the information provided by the ClinGen Expert Panels, we can avoid using criteria that are no longer appropriate. Next, we will look at the criterion PS4, which was scored at a moderate evidence level by the ClinGen expert panel.
If we look at the comment for this criterion, we can see that this recommendation was based on two probands that met the RUNX1 phenotype criteria, demonstrating an increased prevalence in affected individuals. We have also been provided the PubMed ID, allowing us to review the referenced publication and validate the claims made by the ClinGen expert panel. By incorporating ClinGen Expert Curated Interpretations, VSClinical can streamline the variant interpretation process while providing maximum transparency through the inclusion of criteria comments that provide detailed explanations for all recommended criteria.
IARC TP53 Database
In 2016, the International Agency for Research on Cancer published the IARC TP53 Database, which compiles occurrence and phenotype data on TP53 germline and somatic variants linked to human cancer. While many variants in TP53 would be classified as variants of uncertain significance if evaluated using the original ACMG guidelines, by incorporating data from the IARC TP53 database, the impact of these variants can be better understood. In December of 2020, the ClinGen TP53 Expert Panel and other researchers and institutions published a new set of ACMG specifications adjusting the rules for applying the ACMG criteria to variants in TP53. These criteria modifications include updates to the allele frequency thresholds for BA1 and BS1, an adjusted point system for PS2 and PM6, and recommendations for modifying the strength of the PP1 criterion, allowing it to be applied at a strong evidence level. The following image shows a decision tree that specifies how the PS3/BS3 criteria have been modified for TP53 variants:
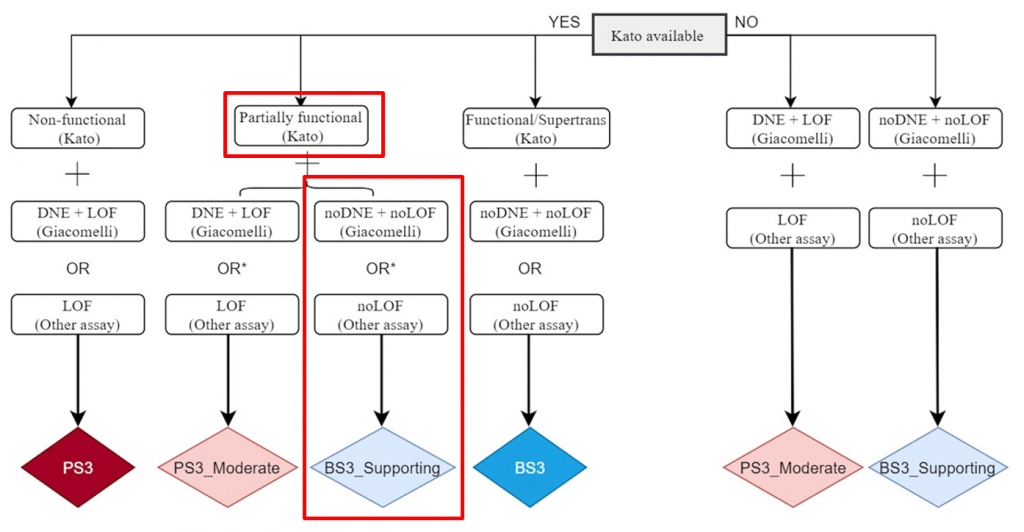
The first branch in this decision tree uses the 2003 transactivation assay published by Kato et al. to determine if the variant results in a low functioning allele. The next branch of this decision tree uses data published by Giacomelli et al. to assess whether there is evidence of a dominant-negative effect or evidence supporting loss of function. If no other assay supports a loss of function, then criteria BS3 is applied at a supporting level of evidence.
As you can imagine, manually assessing each variant using this decision tree can be both time-consuming and error-prone. Thankfully, this decision tree, along with all other parts of the TP53 classification process, can now be automated using VSClinical’s Previously Interpreted Variant Sources feature in conjunction with the IARC TP53 Database. To add this feature to your workflow, simply open the ACMG Project Options, navigate to the Previously Interpreted Variant Sources tab, and use the dropdown to add the TP53 ACMG R20, IARC track.
Once the IARC TP53 track has been added, we are ready to begin evaluating our variant. For this example, we will look at the variant TP53 c.422G>A, a missense mutation in exon 5 of the NM_000546.6 transcript. Looking at the variant tab, we can see that several criteria have been automatically applied, and we have a recommended classification of likely pathogenic.
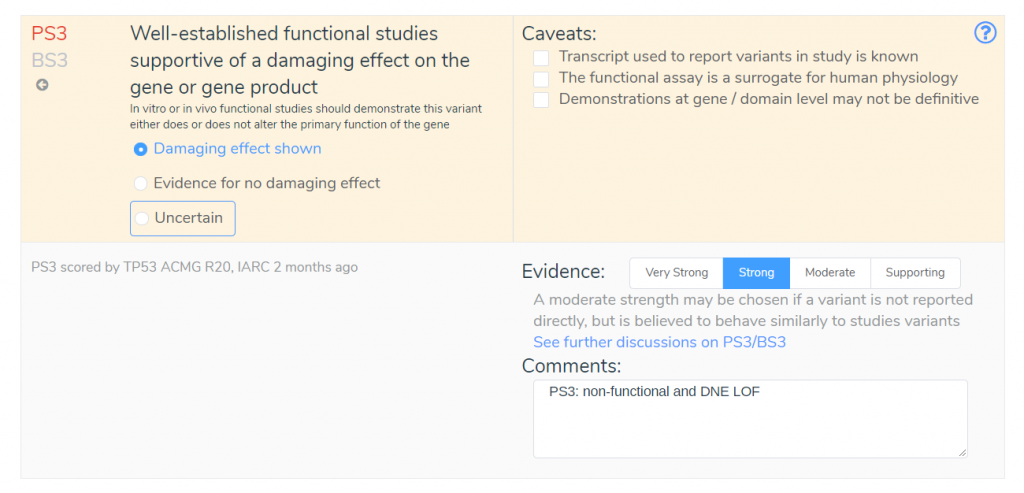
Notice that PS1 is not selected, even though it is recommended by the VSClinical recommendation engine. If we examine the TP53 guidelines published by the ClinGen expert panel, we can see why this criterion was excluded. While this variant is classified as pathogenic in ClinVar, the TP53 guidelines specify that this criterion requires confirmation via RNA that there is no difference in splicing and can only apply to variants classified as pathogenic by the ClinGen TP53 Expert Panel. Our evidence summary shows that PS3 has been applied instead of PS1, indicating that functional studies support a damaging effect on the gene product.
By looking at the comments associated with PS3, we can see that this criterion has been applied due to evidence demonstrating a low functioning allele and evidence of a dominant-negative effect. Without this new IARC TP53 Database, we would be missing this vital information about this variant. We would be forced to manually evaluate the updated TP53 specifications, which could result in an incorrect application of criteria such as PS1.
Conclusion
In this blog post, we demonstrated the utility of incorporating expert-curated variant interpretations into your VSClinical workflows and discussed how this feature can be used to leverage the ClinGen Expert Curated Variant Interpretations track and the new IARS TP53 Database when evaluating variants. We are excited to announce the upcoming release of this new feature which will be available later this year. If there are any questions about the content of this blog post, feel free to leave a comment below. If you would like to see a demo or trial the software, you can click on the button below to request a VarSeq evaluation: