VSPipeline is a command-line interface that will provide high throughput environments the ability to tap the full power of VarSeq’s algorithms and flexible project template system from any command-line context, including the existing bioinformatics pipeline. This feature is a great resource for analyzing large sample volumes as it automates importing and annotating your data, which can help streamline your analysis time. One of our most recent updates to VSPipeline is the ability to import externally called CNVs. As of VarSeq 2.2.3, VSPipeline can now import externally called CNV files in VCF format for single-sample and multi-sample projects.
The foundation of VSPipeline is based on a project template that defines the workflow of the analysis. In this example, the project template utilizes the ACMG sample classifier, which evaluates variants according to the ACMG guidelines, and a table specific to imported CNVs. Once the template is saved, going to File>Save Project as Template can then be defined in the VSPipeline script. As highlighted below, the template is called “ACMG Workflow.”

The next requirement is to define the samples to be imported into the template and the sample manifest text file. The manifest text file can automatically import sample and patient information and define the BAM file paths to the samples of interest. An easy way to get the correct layout for the manifest text file is to export the samples table of an existing project in VarSeq; an example is shown in Figure 2.

Once the manifest text file is defined, the next step is associating the CNV VCF file with the samples of interest. If the sample names match, the corresponding cnv.vcf file will import and associate with the correct sample in the project. For example, this script specifies that SAMPLE1 has an associated CNV file called SAMPLE1.cnv.vcf.gz, Figure 3. This option also indicates the tableID, Table1, for the CNV file to be imported into the project.

After defining the template and required files, the last step is running the script. VSPipeline can be accessed through the command line terminal or through VarSeq by going to Tools>Open Folder>Program Folder and selecting VSpipeline. This will open up the pipeline terminal, from which the following command can be entered: batch file=” pipeline script.” As shown in Figure 4, the name of the example pipeline script is VS_Pipe_cnv_import_example. Once the script is initiated, the output will create a project with all specifications, including a table with externally called CNVs, Figure 5.

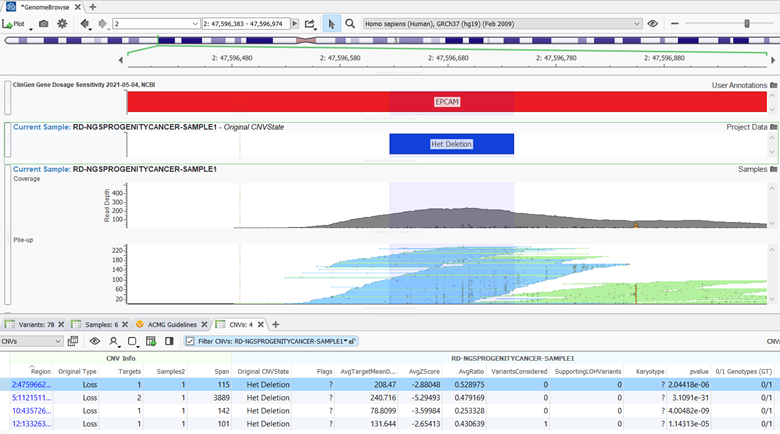
Together VSPipeline can be implemented to automate your NGS pipeline solution and is valuable when analyzing large sample volumes. As discussed, this feature now supports the ability to import external CNVs in a VCF file, and an example of the script can be found in the documentation. If you would like to see a more detailed presentation on this feature, please watch one of our most recent webcasts https://www.goldenhelix.com/resources/webcasts/vsclinical-a-complete-clinical-solution/index.html. Additionally, if you would like to have a trial of VSPipeline, please reach out to info@goldenhelix.com.