ICGC’s database is now available
For quite a while, COSMIC has been synonymous with the catalog of “known somatic mutations”. It is the ClinVar of cancer mutations and invests heavily in “expert curation” (having human experts read papers and pull out references to published somatic mutations).
But it turns out there is a new kid on the block, and he is huge: ICGC or the International Cancer Genome Consortium.
How huge? 1 billion dollars and over ten years. That’s how huge. (See this fantastic recent interview with Francis Ouellette on PLOS for more mind-boggling stats).
ICGC and the encompassing The Cancer Genome Atlas (TCGA) projects are very bold and clear in their goal: to capture the full spectrum of mutations that occur in cancer and their effects in transcriptional regulation.
Started in 2008, and planning to run until 2018, they aim to cover 500 different tumor types, each with 500 matched tumor/normal samples for each type. That’s 50,000 total genomes!
Note the 1000 Genomes Project, the previous largest public genome sequencing effort, made it to 2,500 genomes.
Well, how are they doing? Pretty darn well! If you go to the ICGC data portal you will see a list of all of the cancer genome projects that have submitted data to date.
At this moment, they have ~17,000 donors (which remember provide both a tumor and a normal sample), ~9,000 of which have had sequencing performed and somatic variants called.
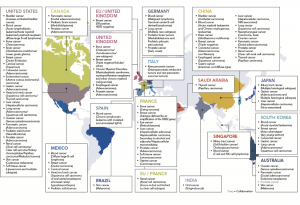
Here is a map of individual projects of this international effort and the tumor types they contribute.
As these projects continue to contribute their data, the ICGC composes unified variant catalog releases 3-4 times a year in versioned data releases.
The latest, v20, has 36,985,985 simple somatic mutations in 57,637 mutated genes!
To compare that to some of the other variant catalogs (mostly germline mind you):
- dbSNP 144 – 144,897,296
- 1KG Phase 3 v5b – 85,823,495 from 2,504 individuals
- ExAC 0.3 – 10,324,246 from 61,486 individuals
- COSMIC 71 – 2,151,007 (After v71, they started bulk-importing TCGA variants)
- ClinVar 2016-04-05 – 144,905
Visualize, Annotate, Interpret
While you can go to the ICGC data portal and download a VCF with heavily delimited text blob INFO fields for each of these 36 million variants, it takes a lot of cross referencing and eye squinting to make much of it.
Once de-parsed, what you get is a per-project detailed breakdown of the number of donors that had the variant versus the total number of donors and this ratio as an “Affected Donor Frequency”.
It took us a while to settle on the most useful way to transform and represent all this data in a visually intuitive and source-representative way while making the data very useful for bulk annotation and filtering.
In the end, I think we nailed it.
Here are some screen shots our curated “Simple Somatic Mutations 20, ICGC” track in our GenomeBrowse visualization (available in stand-alone GenomeBrowse, or part of our analysis tools VarSeq or SVS).
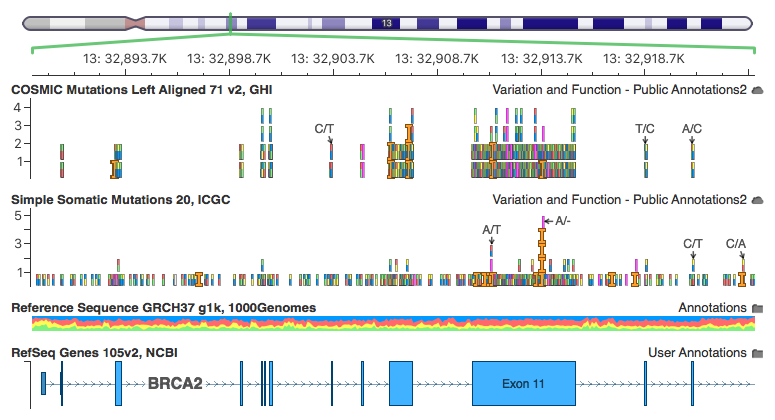
Part of BRCA2 comparing somatic mutations between COSMIC and ICGC. Note ICGC covers many intronic variants, while COSMIC mostly contains exonic.
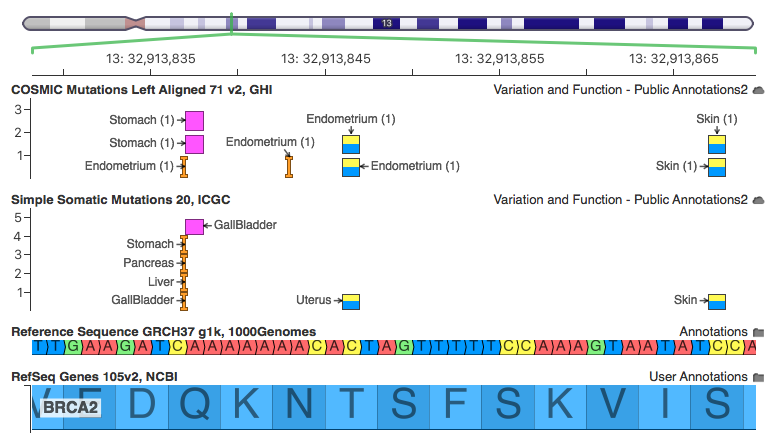
This region has a number of rare variants (occurring in only 1 sample in COSMIC and a handful in ICGC).
The insertion on the left has one record for each project that observed it. This is the record for the PAPC-AU project with a primary site of the Pancreas. Note that 2 samples for this project have the mutation out of 392, but 5 samples across all projects have this mutation (the other 3 in the visualization all have 1 affected donor each).
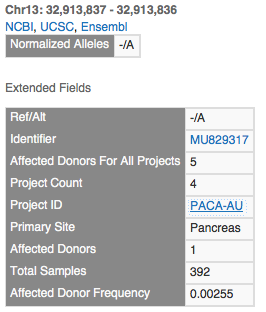
The available fields in ICGC, including useful hyperlinks back to the ICGC data portal at the variant and project level.
Cancer Interpretation
While COSMIC’s expert curation method provides rich literature references, ICGC clearly leads in the ability to profile the prevalence and types of cancers a given mutation occurs and will no doubt become an indispensable tool in the interpretation of somatic mutations in clinical genomics.
We plan to keep our public annotation library updated with the latest ICGC release (and of course, keeping access for all historical ones). Go ahead and check it out, and let us know what you think!