Use the force of evaluation scripts to automate and customize your VSClinical ACMG workflow in VarSeq 2.4.0. VarSeq 2.3.0 came packed with new features! Most notably, VarSeq variant analysis expanded to support the import and annotation of structural variant files, and the AMP cancer workflow in VSClinical gained new functionality with the addition of evaluation scripts which help automate and… Read more »
Discover the power of VSClinical’s Interpretation Match Behavior options for managing the scope of somatic variants in cancer reporting, enabling clinical teams to make informed treatment decisions. Multiple interpretations can apply to a single biomarker or tumor type. In some circumstances, a clinical team may only want to report the most relevant and significant biomarker, treatment, diagnosis, or prognosis interpretations… Read more »
Explore the cutting-edge capabilities of VarSeq in prenatal genetic screening as we delve into real-life cases, expert analysis, and efficient strategies to quickly assess Whole Exome Sequencing (WES) samples for genetic abnormalities in our recent webinar. Thank you all to those who attended our VarSeq webinar covering prenatal genetic screening! We had a great turnout and loved hearing from our… Read more »
Discover the innovative clinical evidence search of the VSClinical AMP workflow, where patient biomarkers and tumor types are matched to the most relevant data from top sources like CIViC, DrugBank, and Clinical Trials. One of the core features of the VSClinical AMP interpretation workflow is the ability to search third-party data sources to find clinical evidence that matches the patient’s… Read more »
VSClinical AMP Matching of Interpretations In this blog post, we will delve into the intricacies of the VSClinical AMP interpretation workflow. At the heart of this process lies the task of annotating cancer biomarkers with the correct interpretations based on the classification of the tumor and the type and scope of the biomarker. This is a crucial step in understanding… Read more »
Oftentimes, the endpoint of a clinical variant analysis is a standardized, clinical report. As such, we ship a number of default templates with VSClinical for users to report their findings. But these templates are just a starting point! Our platform allows users to fully customize their reports to adhere to lab-specific preferences. We have shared a plethora of how-to’s on… Read more »
Thanks to all those who attended the recent webcast by Dr. Rana Smalling, “Integrating Custom Gene Panels for Variant Annotations”. If you were unable to attend or would like to recap, here is a link to watch the broadcast. We covered a lot of content regarding virtual gene panels, and there were several questions submitted during our Q&A session that… Read more »
In our previous blog, we covered the highlights of our Advanced Report Customization in VSClinical webcast in the context of germline clinical reports. Now, we bring you the next of the series: somatic clinical reports. In the recent webcast, Advanced Report Customization, we covered a range of somatic-focused clinical reports, demonstrating how easy it is to create AMP guideline-based clinical… Read more »
In February 2020, the American College of Medical Genetics (ACMG) and the Clinical Genome Resource (ClinGen) published a joint consensus on standards for the interpretation and reporting of copy number variants (CNVs) ranging from large CNVs spanning multiple genes to small intragenic events1. The guidelines consist of over 80 different criteria which are arranged into five distinct sections. These extensive… Read more »
As a newer member of the Golden Helix FAS Team I, like many of our customers, went through a phase of learning the ins, outs, and shortcuts of clicking around the program. In an effort to expedite new users coming up to speed, or to teach some tricks to experienced users, I’ve compiled a list of the top things I… Read more »
In order to thoroughly assess a variant’s pathogenicity, it is important to take into account the variant’s effect on splicing. While the interpretation of variants that disrupt the pairs of bases at the beginning of a splice site is fairly straightforward, variants resulting in the introduction of a novel splice site are more difficult to interpret. In this blog post,… Read more »
One of the many tricks of encoding so much functionality into so little space in eukaryotic genomes is the ability to produce multiple distinct mRNAs (transcripts) from a single gene. While one transcript is often the dominant one for a given tissue or cell type, there are, of course, exceptions in the messy reality of biology. It doesn’t take many… Read more »
Thank you to those who attended the recent webcast, “Exome Analysis with VS-CNV & VSClinical: Updated Strategies & Expanded Capabilities”. For those who could not attend but wish to watch, here is a link to the recording. In this webcast, we covered the capabilities and updates that have been incorporated into VarSeq that enhance whole exome sequencing workflows. The new… Read more »
Clinical testing labs produce reports as the end product of the NGS variant detection and interpretation workflow. Necessarily, the content, detail, and presentation of the report needs to be specialized to each clinical lab, and potentially each offered test. Our last blog post introduced the new Word-based report templates in VSClinical. In this blog post, we will introduce and explore… Read more »
The collaboration between the Clinical Genome Resource (ClinGen) consortium and the American College of Medical Genetics (ACMG) recently developed published guidelines for the interpretation of CNVs called on next-generation sequencing data. These new guidelines are the first to provide a robust set of rules for the interpretation of small intragenic deletions and duplications and are now automated in VSClinical. … Read more »
The recent release of VarSeq 2.2.2 brings our Word report template system, previously featured in VSClinical AMP, to the VSClinical ACMG workflow. This blog post will describe how to use the Word template system using one of our shipped templates as well as how to start customizing your own templates. We will cover the three different report templates that ship… Read more »
VarSeq recently received major upgrades in a wide range of areas, one of these areas includes adding annotations such as GnomAD. This includes new fundamental methods of CNV ACMG guideline processing but also a large number of small additions in annotations. One addition is the application of gnomADs – Gene Constraints. This provides various metrics for pathogenicity on a per… Read more »
Annotating genomic variants is a very complex process but perhaps the most important part of next-generation sequencing variant analysis. Here at Golden Helix, we recognize the importance and value of having the most up-to-date sources available and curating new annotation sources as they become available for variant analysis. Golden Helix has curated over 100 annotation sources for human variant analysis… Read more »
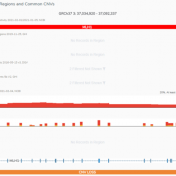
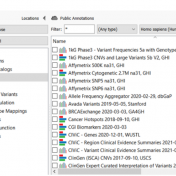
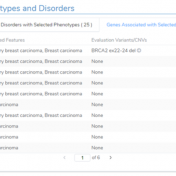
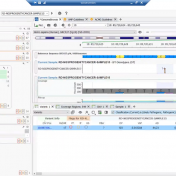
Our recent release of VarSeq 2.2.2 comes with a long list of upgrades and new features. In this blog post, we will demonstrate how defining sample phenotypes are available in VSClinical. One noticeable change is the ACMG guideline variant evaluation in VSClinical. Not only has this interface added CNV guideline evaluation, simplified the reporting process with embedded Microsoft Word and… Read more »
Didn’t catch the webcast live? No worries! We cover ‘VSClinical: A Complete Clinical Workflow Solution’ Q&A’s in this blog post. The webcast, ‘VSClinical: A Complete Clinical Workflow Solution’ demonstrated how solutions provided by Golden Helix can be implemented to cover all requirements of a clinical workspace. Specifically, this webcast focused on a detailed workflow from a bioinformatician, geneticist, and lab… Read more »