September 13, 2017 12:00 PM, EST This month we’re branching out and covering a topic we’ve never explored before – annotation capabilities! Darby Kammeraad, Field Application Scientist at Golden Helix, will be giving us some insight into the advantages of VarSeq’s capability with annotations. The number of annotation topics to cover are seemingly limitless. In this webcast, we will focus… Read more »
We don’t just like hearing what our clients are up to … we love bragging about what they’re doing to the world as well! This week we’re showcasing Dr. Stanley Nelson, and his team at UCLA Health, who used next-generation sequencing and our VarSeq software to help diagnose a child’s long running medical mystery. Audrey Lapidus knew there was something… Read more »
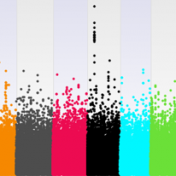
We are dedicating our August webcast to explain the latest and greatest enhancements with GWAS workflows in SVS. Below you can find further details and registration information. We hope you can attend! Wednesday, August 9th, 2017 12:00 PM EST In this webcast, we will focus on the recent improvements to our research product SNP & Variation Suite. Over the past… Read more »
We’re gearing up for ASHG and kicking off our annual T-Shirt Design Competition! This one goes out to all the clever, witty genomic jokesters out there … here’s your chance to cash in on those DNA punchlines you’ve been practicing! Here’s how the T-Shirt Design Competition works: Draw it out or write it down; we accept either illustrated or written designs. Email… Read more »
Our July webcast presentation will be focused on clinical workflows in VarSeq. We wanted to share the full details with you and hope you are able to attend! Wednesday, July 12th 12:00 PM EST This month’s webcast is a VarSeq exploration, featuring several example workflows and helpful features in VarSeq that can be used in the clinic. We will discuss… Read more »
When studying complex diseases that may have multi-genic contributions from across the genome, it is not uncommon to see what may appear like elevated correlation between your trait or other test variable and the SNPs across the genome. The problem is at first glance you won’t be able to tell if this is due to a population structure in your… Read more »
We have been heads down doing the detailed and careful work to improve our CNV caller algorithm in the past three months since our we launched our Exome capable CNV caller and are very excited about the massive step forward we have made with the VarSeq 1.4.5 release. Additionally, we have added the all new Whole Genome large-event caller capable… Read more »
Low read depth? Great! We are excited to introduce our new CNV calling algorithm for low and ultra-low read depth Whole Genome Sequencing (WGS) data. This algorithm is designed to call large cytogenetic events with high confidence from low read depth whole genome data, with as few as one million aligned reads or 0.02x coverage. The low sequencing cost of… Read more »
We are ending the first half of the year with a webcast featuring our entire clinical software stack: Golden Helix’s End-to-End Solution for Clinical Labs. This webcast is scheduled for Wednesday, June 7th at noon EST, and it’s not one you’ll want to miss! Here are the full details: Wednesday, June 7th 12:00 pm EST In this webcast we will provide an… Read more »
In just a few days, Golden Helix will arrive in Copenhagen, Denmark to attend the 2017 European Society of Human Genetics Conference (ESHG). We are excited to be returning to ESHG for the second year in a row! It’s always a pleasure to see our European customers and meet some new faces in the community. This year the Golden Helix team will be… Read more »
We are excited to announce our next webcast, The Sentieon Genomic Tools – Improved Best Practices Pipelines for Analysis of Germline and Tumor-Normal Samples. This webcast is scheduled for Wednesday, May 17th at noon EST and will focus on our new partnership with Sentieon. Wednesday, May 17th 12:00 pm EST With next-generation sequence datasets frequently reaching petabytes in size, processing genomic data accurately… Read more »
Today, we are happy to announce a multi-year partnership with Sentieon, a company that develops bioinformatics secondary analysis tools to process genomic data. This partnership will integrate Sentieon’s secondary analysis tools with Golden Helix software to provide users with a comprehensive solution for genomic data analysis. Sentieon’s suite of secondary analysis tools made the significant improvement in runtime over BWA-MEM, GATK,… Read more »
We are pleased to announce our next webcast, Identifying genetic variants associated with rare Mendelian Diseases. The live event is is scheduled for Wednesday, May 3rd at noon EST. Here are the specifics: Wednesday, May 3rd 12:00 pm EST Dr. Jingga Inlora is a postdoctoral fellow in the Snyder lab at Stanford University. In this webcast, Inlora will present on… Read more »
Recently, we added a natively supported Genotype Phasing and Imputation capability in SNP & Variation Suite 8.7.0. Since then we have had fantastic feedback and adoption as folks take advantage of the BEAGLE 4.0 and 4.1 algorithms from within their existing SNP GWAS and agrigenomic workflows. One piece of feedback we heard from our time at PAG, ACMG and our… Read more »
We are pleased to announce our next webcast, CNV Analysis in VarSeq – A User’s Perspective. The live event is is scheduled for Wednesday, April 19th at noon EST. Here are the specifics: Wednesday, April 19th 12:00 pm EST Clinical labs must have the ability to go from a collection of samples to a professional report documenting a short list… Read more »
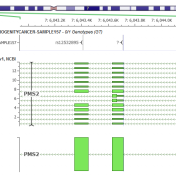
It may be possible to say that annotating a variant correctly and accurately against gene transcripts is the most important job of a variant annotation and interpretation tool. We take it very seriously at Golden Helix as we support VarSeq and its use by our customers in both research and clinical contexts. It has been a source of frustration that… Read more »
Join our upcoming webcast : Wednesday, April 5th 12:00 pm EST Dr. Reza Sailani is a Research Fellow in the Genetics department at Stanford University. To provide an overview of his research, Sailani will present on the following two recent studies he has conducted: Association of AHSG with alopecia and mental retardation (APMR) syndrome: Alopecia with mental retardation syndrome (APMR) is… Read more »
Today, I am happy to announce the introduction of VS-CNV for Gene Panels and Exomes. We have developed this capability in partnership with PreventionGenetics. PreventionGenetics will use VarSeq CNV for analysis of gene panels, and in the future for exome sequencing. The software gives PreventionGenetics the opportunity to conduct a comprehensive CNV analysis on NGS data, in many cases eliminating the need… Read more »
ACMG 2017 is just around the corner! We are halfway through March already and it’s just about time to head off to sunny and warm Phoenix, Arizona. While the temps have been mostly mild for the last few weeks in Montana, I bet those of you in the northeast are looking forward to your time in Phoenix! You will find… Read more »
Ever since the MacArther Lab announced the new gnomAD browser at last year’s ASHG conference, we have had many requests from our customers to make this new variant frequency source available within both VarSeq and SVS. This new dataset includes variants obtained from 123,136 exome sequences and 15,496 whole-genome sequences. In comparison to the original ExAC dataset which contained exomes… Read more »