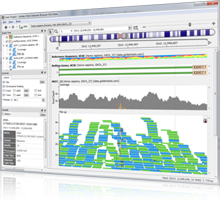
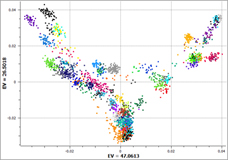
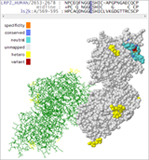
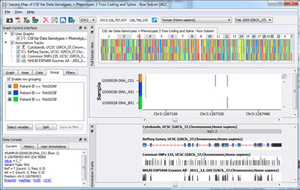
Presenter: Dr. Bryce Christensen, Statistical Geneticist Date: July 24, 2013 Duration: 60 Minutes Abstract Golden Helix GenomeBrowse™, a free visualization tool for all types of sequence data, was introduced in 2012 to broad acclaim. Researchers using GenomeBrowse discovered a product far beyond the status quo with seamless navigation of sequence alignments and other genomic data using a fluid, fast, and… Read more »
In a couple of short weeks, Gabe is headed off to TCGC in San Francisco where he will be giving part of a short course. He was super excited about it last year and is even more so this year. I sat down with him yesterday to find out why. Jessica: What’s TCGC? Gabe: Last year I got to attend… Read more »
Last month, Bryce Christensen wrote a little about mixed models and their application in GWAS. He promised that this analysis would be available in SNP & Variation Suite (SVS) “soon,” but didn’t elaborate. We are now excited to announce that three mixed model methods are available in SVS: GBLUP, EMMAX, and MLMM! To help demonstrate their utility and when to… Read more »
Presenter: Greta Linse Peterson, Senior Statistician Date: Wednesday, June 5th, 2013 Time: 12:00 pm EDT, 60 minutes Abstract Population structure and inbreeding can confound results from a standard genome-wide association test. Accounting for the random effect of relatedness can lead to lower false discovery rates and identify the causative markers without over-correcting and dampening the true signal. This presentation will… Read more »
Presenter: Dr. Bryce Christensen, Statistical Geneticist and Director of Services Date: Wednesday, May 15th, 2013 Time: 12:00 pm EDT Abstract Next-Generation Sequencing analysis workflows typically lead to a list of candidate variants that may or may not be associated with the phenotype of interest. Any given analysis may result in tens, hundreds, or even thousands of genetic variants which must… Read more »
Presenter: Khanh-Nhat Tran-Viet, MHA, Manager/Research Analyst II at Duke University Date: March 7, 2013 Time: 12:00 pm EST, 60 Minutes Abstract Technological advances in next generation sequencing provide clinicians and researchers with more effective methods to identify pathogenic gene mutations for heritable diseases. To date, the National Eye Institute Bank lists over 450 genes associated with eye-related disorders. Analytical processing… Read more »
Twenty-four new variants discovered, each conferring more than a 2-fold risk of developing ASD Date: January 29, 2013, Noon EST, 90 minutes Presenters: Dr. Hakon Hakonarson, Director of the Center for Applied Genomics, Children’s Hospital of Philadelphia Dr. Mark F. Leppert, Professor of Human Genetics at the University of Utah and Chief Science Advisor at Lineagen Dr. Michael Paul, President… Read more »
Join me on December 5th for a one-hour webcast as I explore my personal exome provided by the Exome Pilot project of 23andMe. Exome sequencing has seen many success stories in the realm of diagnosing highly penetrant monogenic disorders as well as in informing treatment of certain cancers. As the use of exome sequencing expands to more complex polygenic disorders… Read more »
After much anticipation, we are excited to announce that SVS 7.7 is available! It’s been a lot of work, but we are pleased with the result and know you will be too! SVS 7.7 includes: Improved RNA-Seq Analysis Package for mRNA Expression Profiling Import tabularized quantification data from the Golden Helix pipeline or other sources. Utilize DESeq to perform differential… Read more »
Golden Helix is proud to announce our participation in EA’s 2012 “Leave Your Fingerprint on the Cure.” This event will be taking place at ASHG 2012 in San Francisco at the Moscone Center on November 7th. The fifth annual “Leave Your Fingerprint on the Cure” was designed by EA to help raise money for researchers in their efforts to discover… Read more »
Date: Wednesday, October 24th, 12:00 pm EDT | 4:00 pm UTC Presenter: Gabe Rudy, VP of Product Development Abstract Next-generation sequencing of DNA has shown to be a successful tool to discover causal variants of rare, highly penetrant mendelian diseases. In this webinar, we will analyze clinical samples to demonstrate the bioinformatic workflows and visualization techniques used to reproduce the… Read more »
Golden Helix announced it would offer a complimentary genome browser for visualizing DNA and RNA-Seq data back in October of 2011; however no release date was disclosed at that time. Well, after much blood, sweat, and tears, we are VERY excited to announce that GenomeBrowseTM is here! GenomeBrowse raises the bar on the on the experience of exploring and finding… Read more »
As first announced in October 2011 and previewed in a blog post last month, Golden Helix has been hard at work for the last year developing a free genome browser called GenomeBrowseTM. We are now beyond excited to announce that GenomeBrowse will be available to the public on September 12th on our website. (Did we mention it’s free?) GenomeBrowse raises… Read more »
Thank you to everyone who joined us yesterday for a webcast by Dr. Ken Kaufman of Cincinnati Children’s Hospital: “Identification of Candidate Functional Polymorphism Using Trio Family Whole Exome DNA Data.” Over 750 people registered for this event and 430 attended – a new Golden Helix record! If you missed the webcast (or would like to watch it again), the… Read more »
Recently I gave a presentation on bioinformatic filtering: the process of using quality scores, annotation databases, and functional prediction scores to intelligently and quickly reduce your variant search space. View the recording here» In this webcast, I mention that filtering is something we have been doing for a long time, and that there are some great examples that use exome… Read more »
Bioinformatics is a serious thing. Petabytes of data that hold the truth to the genetic underpinnings of the human species, among many others, are generated and analyzed by the world’s leading scientists every year. Here at Golden Helix, we thought that there was plenty of room to add a little humor into the bioinformatics world. Meet Dr. X. He is… Read more »
It’s that time again! We here at Golden Helix are excited to announce SVS 7.6 with more features for DNA-Seq analysis, the addition of RNA-Seq functionality, reorganization of SVS into “packages” including two new ones, and the release of new plot types enabled by Matplotlib. It’s certainly been busy as we pack all this into the sixth installment of the… Read more »
Recently we have expanded our annotation track offerings with new human variant frequency catalogs such as the 1000 Genomes Phase 1 Data. Of course, we also curate data for plant and animal genomes – some of which are currently available in our software and some of which will be available in our next release. In this blog post, I will… Read more »
I can’t believe it’s that time again! Next week we are heading off to Montreal, Canada for the week of October 11-15 for the 2011 American Society of Human Genetics annual meeting, this year in conjunction with the 12th International Congress of Human Genetics. We are excited as ever to hear what everyone has been up to, how your studies… Read more »
More powerful rare variant analysis and visualization techniques We are proud to unveil SNP & Variation Suite (SVS) 7.5! The fifth installment of SVS delivers the most powerful rare variant analysis tools with the latest advances in tertiary or “sense making” methods for DNA next-gen sequencing. This release also adds some cool and productive features to our popular Genome Browser…. Read more »