Several of our customers have published recently, using the SVS software and I wanted to share their work. Congrats to all! Daria Babushok and colleagues at The Children’s Hospital of Philadelphia published Emergence of Clonal Hematopoiesis in the Majority of Patients with Acquired Aplastic Anemia in the Cancer Genetics Journal which used comparative whole exome sequencing to evaluate clonal hematopoiesis in acquired aplastic… Read more »
A few of our customers have published recently and I would like to take the time to both recognize them for their achievement and pass on their articles. Enjoy! Kazima Bulayeva at the Vavilov Institute of General Genetics and her colleagues recently published Genomic Structural variants are linked with intellectual disability in the Journal of Neural Transmission. The paper looks at mutations… Read more »
Last month, Dr. Bryce Christensen presented Population-Based DNA Variant Analysis via webcast. The webcast reviewed the fundamentals of population-based variant analysis and demonstrated some of the tools available in SVS for analysis of both common and rare variants such as the SKAT-O method, as well as other functions for annotation, visualization, quality control and statistical analysis of DNA sequence variants. Here… Read more »
Today, Golden Helix and Fluxion Biosciences announced a collaboration in a value-added reseller relationship. The relationship will bring the VarSeq software application to Fluxion’s global client base providing them with a method to study tumor DNA in the circulation. Fluxion is proud to offer the capability as it helps move them toward their goal of offering a complete sample-to-answer workflow for… Read more »
Our Genomic Prediction webcast in December discussed using Bayes-C pi and Genomic Best Linear Unbiased Predictors (GBLUP) to predict phenotypic traits from genotypes in order to identify the plants or animals with the best breeding potential for desirable traits. The webcast generated a lot of good questions as our webcasts generally do. I decided to begin to share these Q&A… Read more »
In just 5 days, the 22nd International Molecular Medicine Tri-Conference (Tri-Con) will kick off in San Francisco. This year, Tri-Con will offer over 3,000 attendees 6 symposia, over 20 short courses, and 17 conference programs focused on drug discovery, genomics, diagnostics, and information technology surrounding, molecular medicine. Both Dr. Andreas Scherer, CEO of Golden Helix and Gabe Rudy, our Vice President of… Read more »
Today, we at Golden Helix announced our collaboration with PreventionGenetics as they prepare to implement the VarSeq software into their exome sequencing pipeline. The VarSeq software will allow PreventionGenetics to offer an exome test by dramatically speeding up the analysis process. VarSeq will narrow down sequence data into gene(s) of interest based on inheritance patterns, facilitating the identification of clinically relevant… Read more »
Genotype imputation is a statistical technique for estimating sample genotypes at loci that were not directly assayed by sequencing or microarray experiments. There are several reasons why you might want to use imputation in a research study. For example: Improve call rates in GWAS by imputing sporadic missing genotypes Harmonize the data content from different GWAS genotyping platforms so that… Read more »
With January officially in the bag, 2015 is off to a great start, especially for some of our customers who have recently published. I wanted to take a minute to share them with you. Sander van der Laan at University Medical Center Utrecht, published Variants in ALOX5, ALOX5AP and LTA4H are not associated with atherosclerotic plaque phenotypes: The Athero-Express Genomics Study which assessed the impact of common variants… Read more »
In recent months we have been updating our public annotation library to include the most recent versions of existing sources, as well as include new sources. All of these annotation sources are compatible with our three major products, VarSeq, SVS, and GenomeBrowse, and can be used for visualization, annotation, and filtering. dbNSFP NS Functional Predictions 2.8, GHI and dbNSFP Predictions… Read more »
One frequent question I hear from SVS customers is whether whole exome sequence data can be used for principal components analysis (PCA) and other applications in population genetics. The answer is, “yes, but you need to be cautious.” What does cautious mean? Let’s take a look at the 1000 Genomes project for some examples.
The Integrative Therapies Institute is soon hosting the annual, ITI 2015 conference January 23rd through the 25th in sunny San Diego and our own Dr. Andreas Scherer has been invited to speak. Some of the most prominent genomic and integrative medicine specialists will gather at ITI 2015 to share case studies and protocols with the community. Attendees can expect to… Read more »
Once again, we will be kicking off our year with our annual trip to San Diego for PAG XXIII. This year, it could not come at a better time. Over the last few weeks, it has been bitter cold in Montana with temps barely reaching above zero degrees and I for one am looking forward to the warm sun. And… Read more »
If you’ve seen the recent webinars given by Gabe Rudy and Bryce Christensen, you’ve no doubt been impressed by the capabilities of VarSeq when it comes to annotation and filtering. However, we sometimes forget that the power that enables all this complex analysis can also be used in more mundane tasks like VCF subsetting. And although these day-to-day tasks don’t… Read more »
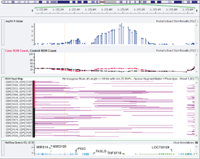
It’s come to my attention in recent weeks, through various customer interactions, that many are not aware of the fantastic functionalities that exist in SNP and Variation Suite (SVS) for large-n DNASeq workflows; this includes large cohort analyses with case/control variables. The data you’ll see below is the publically available 1kG Phase 1 v3 Exome sequences from 1,092 individuals with… Read more »
A helpful tool that is included in SVS, but many of our customers may not know about, is the ability to create Box Plots or box-and-whisker plots. These are effective visualizations for comparing groups of numerical data through the data quartiles. I’ll take you through a couple different cases with examples.
Recently, I have been thinking a lot about Human Genome Variation Society (HGVS) notation — you know “G dot”, “P dot”, and “C dot”. HGVS has quickly become one of the most common ways to represent variants. It’s no wonder that HGVS nomenclature is used so widely. It provides an easily readable, compact representation of a variant. Since it is… Read more »
Late last month I had the opportunity to attend one of my favorite events: the annual meeting of the International Genetic Epidemiology Society (IGES). This year’s conference was held in Vienna, in conjunction with the Genetic Analysis Workshop (GAW) and the International Society for Clinical Biostatistics (ICBS). The program at IGES this year was very diverse, with content ranging from… Read more »
SVS offers options for performing many different QC functions on genomic data. This blog takes you through some of the most commonly applied filters for various analysis types. Filters for GWAS data vary depending on the type of association tests you are performing. A typical GWAS for a common variant usually requires filters to remove problematic or poorly called variants,… Read more »
Golden Helix is excited to host a webinar on Tuesday August 26th discussing the Genomic Prediction methods which were recently integrated into the SVS software. Genomic prediction uses several pieces of information when calculating its results. Genetic information is used to predict the phenotype or trait for the individuals. The phenotypic trait data can be provided for a subset or for all… Read more »