About Darby Kammeraad
Darby Kammeraad is the Director of Field Application Services at Golden Helix, joining the team in April of 2017. Darby graduated in 2016 with a master’s degree in Plant Sciences from Montana State University, where he also received his bachelor’s degree in Plant Biotechnology. Darby works on customer support and training. When not in the office, Darby is learning how to play guitar, hunting, fishing, snowboarding, traveling or working on a new recipe in the kitchen.
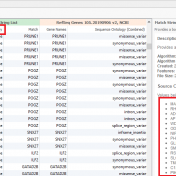
A common discussion with our customers includes the challenges with the tertiary stage of analyzing next-gen sequencing data. This is the stage where all data from gene panels, exome, or whole genome scale pass through filters to quickly isolate the clinically relevant variant contributing to a patient disorder. Golden Helix has recognized these challenges in the scale of data and… Read more »
In this blog update, I’ll be walking you through some of the advanced plotting capabilities with GenomeBrowse. The strategy with any next-gen sequencing analysis is to filter down to interesting variants for either research or clinical conclusion. Golden Helix produces powerful software specifically tailored for this efficient and comprehensive search for interesting and clinically relevant variants. One additional advantage of… Read more »
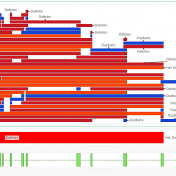
We have now reached the final blog of the NGS-Solutions for Clinical Variant Analysis series. Part I of this series explored the capture of variant classifications in the VSClinical environment when following the ACMG and AMP guidelines. Part II was similar in content but for the capture of clinically relevant copy number variants as well as using a CNV catalog… Read more »
Our software solutions and partners have brought dramatic improvements to the secondary and tertiary analysis stages of variant evaluation. Regarding secondary analysis, we’ve discussed increased efficiencies in speed and overall accuracy in the variant calling process with Sentieon. On the tertiary side, we have explored numerous workflows in VarSeq highlighting filtration to clinically relevant variants, as well as the automated… Read more »
It doesn’t take much effort to find articles discussing the value of Next-Generation Sequencing (NGS). There is a consistent tone amongst authors that implementing NGS pipelines are critical for clinical efficiency in both hereditary disorders and somatic. However, NGS strategies do not come without their own challenges. Challenges include not only the detection and calling of high quality/probability variants from… Read more »
Golden Helix software provides huge analytic gain in handling large-scale genomic data. For example, a number of VarSeq users run cohort projects of whole genome level data processing hundreds of millions of variants at a time. However, many of our users are running gene panel level data for custom panels related to cancer (both hereditary and somatic), autism, cardiac, and… Read more »
Thank you to everyone who joined me for our latest webcast, “Next-Gen Sequencing of the SARS-CoV-2 Virus with Golden Helix.” If you missed the live event and are interested in knowing what we talked about, you may access the recorded event below: Our Live Q&A generated a lot of great questions. Unfortunately, we were unable to answer them all, but… Read more »
Although VarSeq is intentionally designed to be a clinical NGS pipeline tool able to run a handful or even single samples through, we have many users who run large cohort style studies with the tool as well. One common use is to compare case/control data to isolate variants shared among affected individuals and exclude those in unaffected. One incredibly powerful… Read more »
Since the initial release of the copy number variant algorithms in VarSeq, our team has created a variety of content to help users get started with building their copy number variant projects. In our webcast library, you can find a few of our recent webcasts in 2019 covering CNV workflows and validation: Clinical Validation of Copy Number Variants Using the… Read more »
The first two blogs in this series covered Sentieon’s somatic variant calling from Tumor with Normal (Part I) and Tumor without Normal (Part II). In addition to providing multiple somatic variant calling processes, users also have access to high-sensitivity scripts and full support for the GRCh38 reference assembly. Without going into excessive detail, this final blog of the series will… Read more »
To recap what we have covered in this blog series thus far, Sentieon allows users to call somatic variants against a matched normal sample and a tumor-only analysis. Part I of this series covered the variant calling workflow for tumors with matching normal samples. However, a common situation would be to call variants for a tumor sample without the normal…. Read more »
As I mentioned in the first part of this series, Sentieon allows users to call somatic variants against a matched normal sample and a tumor-only analysis. Utilization of a Tumor-Normal Workflow In addition to the fundamental process of alignment and variant calling, there are a few more steps that will improve the quality of your secondary analysis. Figure 1 (below)… Read more »
This month’s webcast delves into VSWarehouse with a focus on our new capability of storing somatic variant projects and catalogs built for the AMP Guidelines within VSClinical. If you didn’t have a chance to join us for the live event, please enjoy the video recording below. Previous webcasts have gone into great detail on the features and processing of somatic… Read more »
Sentieon has become well known for the dramatic improvements it supplies in speed and accuracy for secondary analysis. Luckily, these improvements are not only available for DNAseq germline based variants but also for TNSeq somatic investigation. Sentieon has proven the capability by leading in awards such as the ICGC-TCGA DREAM Challenge for SNVs, Indel, and structural variants. This value not… Read more »
We have covered a lot of ground in this Automating & Standardizing Your Workflows blog series. First, we saw how to perform secondary analysis with Sentieon to generate the necessary VCF and BAM files for tertiary analysis in Part I. The implementation of VSPipeline allowed for rapid import and project generation for a predefined cancer gene panel project template in… Read more »
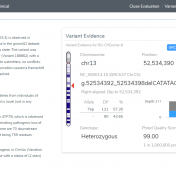
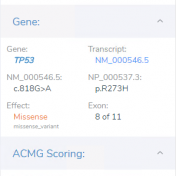
In our last part of this series, we showed how to run a pre-built workflow template via VSPipeline to automatically import and filter sample variants to streamline the search for clinically relevant variants. Now, we can deep-dive into our filtered, pathogenic variants to fully understand and capture their final classification and interpretation. Filtered Germline Variants for ACMG Guidelines The VSPipeline… Read more »
VSPipeline: Automating your Tertiary Workflows The first part of this “Automating & Standardizing your NGS Workflow” blog series covered the secondary analysis steps of read alignment and variant calling with Sentieon. The next step is to transition into the tertiary analysis via utilization of our workflow automation tool, VSPipeline. VSPipeline operates as a command-line tool meant to simplify the deployment… Read more »
Secondary Analysis with Sentieon: Rapid and Accurate Variant Calling This blog post will cover the utilization of secondary analysis tools to produce a list of high-quality variants and associated coverage data. This data will serve as the main, importable content for the tertiary stage of analysis where variants are interpreted and classified for their impact on a patients’ disorder. We… Read more »
As our regular customers may know, we write our blogs to provide relevant content to any NGS-based analysis with VarSeq. Our goal here at Golden Helix is to provide top quality software and guidance on how to use the software efficiently to perform variant analysis. However, this blog series will take a more general perspective and supply some insight into… Read more »
VSClinical provides a rapid-fire way to investigate any variant’s impact by following the ACMG Guidelines process for classification. We will be demonstrating this by looking at interesting examples of rare disorders and showcasing some evaluation steps users may deploy in their analysis. Our first example in this blog series is for a patient who has an indifference to pain, while… Read more »