So, why are we launching a new data warehouse product? Why did we build VSWarehouse? According to Grand View Market Research, the next generation sequencing (NGS) market size was $2.0 billion (USD) globally in 2014. This number is expected to grow from 2015 to 2022 at an annual rate of about 40%. What drives this phenomenon is the increasing number of treatment options for Precision Medicine (see figure 1).
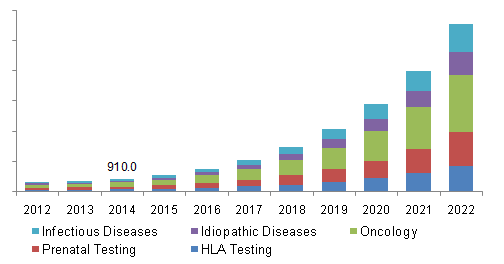 Figure 1: Utility of NGS based diagnostics per disease category
Figure 1: Utility of NGS based diagnostics per disease category
In addition to that, prices of genome sequencing are quickly reducing due to the research & development of rapid, high capacity whole genome sequencers by leading vendors such as Illumina. Obviously, a growing population, increasing desire for prenatal testing, and a growing number of cancer cases in an aging population are expected to further fuel the demand for next generation sequencing based diagnostics.
Now, let’s look at this from a lab’s perspective.
A human genome consists of about 3 billion base pairs. If we were to store all of it we would need about 700 megabytes, assuming that there are no technological flaws to worry about, and therefore no need to include information on data quality along with the sequence. In that case, all we would need is the string of letters A, C, G and T that make up one strand of the human genome, but that is not a valid assumption. This number is actually much higher, about 200 gigabytes, assuming we store all the short reads produced by a sequencer at a 30 x coverage rate. A more compressed representation of this information is the variant called format file (VCF-file), because only about 0.1% of the genome is different among individuals, which equates to about 3 million variants in the average human genome. This means we could derive a “diff file” of just the places where any given individual differs from the normal “reference” genome.
So, per sample we have to store about 3 million variants. Depending on what kind of data we keep, our storage requirements are between 200 megabytes and or 200 gigabytes. The actual amount of data stored per sample depends on the decision how much clinically relevant information we retain (e.g. coverage statistics) as well as how much data we keep for visualization purposes. For example, it would make sense to retain all BAM files associated with a particular sample to display the aligned reads in a Genome Browser. In addition to that, we have to save clinical reports and meta information about the filtering process.
In 2016 labs easily conduct dozens of whole genome analysis per month. Larger labs process hundreds in the same time frame. According to the market study each lab will see on average a 40% increase of its data volume year over year.
This means, very quickly there are billions of variants and terabytes of data to manage per lab. And in a little over two years the newly created data volume will double. In order to capture, organize and leverage this data, advanced warehousing capabilities are required.
The numbers above obviously require a Big Data approach. But the necessity for centralized and genetically aware data management arises already at much smaller variant numbers that are generated by gene panels or more targeted whole exome tests. Essentially, labs require an integrated approach to store, manage and gain insights from samples produced in a clinical setting.
How can you learn more?
- We have a lot of new information about our new product VSWarehouse on our website. Please look here for more information.
- In two weeks, on February 3, 2016, we will have a webcast to introduce VSWarehouse. Gabe Rudy and myself will tell you all about our plans and the product itself that day.
- Finally, around the time of the this webcast, we will lauch my latest e-book on warehousing for genomic data. So, stay tuned.
Thanks for this wonderful information regarding data warehouse.