I’d like to take a moment to announce the release of updated gene tracks for the GRCh38 genome assembly! Gene annotation tracks are essential to all VarSeq projects and workflows. Whether your favorite gene track is Ensembl or RefSeq, both sources have been updated and released and can be used for variant annotation.
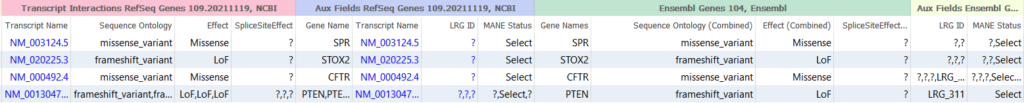
These gene tracks are used to annotate the variant table displaying details about the known transcripts for a gene, the effect of a variant on a given transcript, splice site predictions, and sequence ontology. In addition, much of this data is applied to algorithms used in VSClinical. Of note, the algorithm considers the selection of the clinically relevant transcript for variants of interest. Figure 1 shows the known transcripts for PTEN displayed in VSClinical.

Many components impact this transcript selection. Ultimately, the selection of the clinically relevant transcript for a given gene is completely up to the user and can be saved into a project template for consistency across projects. However, by default the heuristic for preferred transcript selection in VSClinical is as follows:
- Prefer a transcript that is a MANE “Select” transcript
- Prefer a transcript that has an LRG identifier
- Prefer a transcript that has correctly encoded start and stop codons over “incomplete” transcripts
- Prefer a transcript that is protein-coding over one that is non-coding
- Prefer transcripts with longer coding sequences
- If all else is identical, select the first in lexigraphic order
The reason I am going on and on about clinically relevant transcript selection in this blog is that we have updated our Ensembl gene track to include LRG and MANE select transcript fields. Previous versions of RefSeq have already included details on LRG and MANE status but now this data is available for the Ensembl source as well!
If you have any questions about incorporating these sources into your VarSeq projects, or how to change default transcription selection, please feel free to reach out to our support team at support@goldenhelix.com!