Could leveraging CNV analysis for whole exome or genome sequencing help provide answers for the undiagnosed?
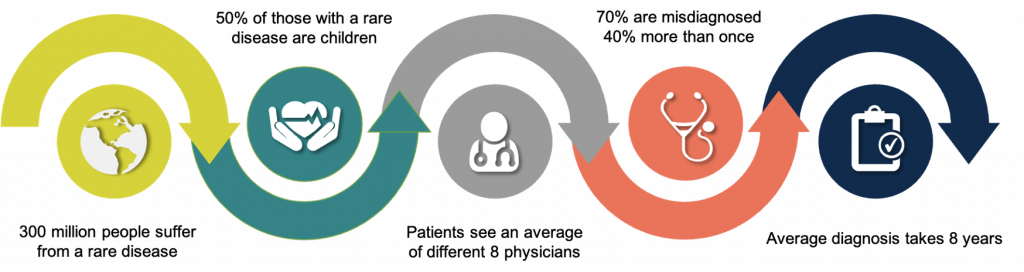
Roughly 4% of the world’s population shares one common desire – a diagnosis (1). Though it may seem counterintuitive, a diagnosis, no matter how grim, can provide relief, validation, the chance to make plans, and have a discernible sense of future for individuals and families whose lives have been put on hold. For an estimated 300 million people suffering from a rare disease, a diagnosis – if one can be found – takes an average of 8 years (2). Many are misdiagnosed during this time. In one study, up to 70% of cases were misdiagnosed, of which 40% were misdiagnosed more than once (3). Of the 6,000 – 7,000 clinically defined rare diseases, 72% are genetic, and of those, 70% present in childhood (1,2). Aptly termed a diagnostic odyssey, the quest for a diagnosis involves an average of 8 different physicians, often forcing people to crisscross the country and undergo a battery of tests. Without an official diagnosis, insurance claims can be refused. Families can quickly find themselves in a financial tailspin, completely inundated by medical bills.
How is it that rare diseases, affecting ~15.5 times more people worldwide than cancer, evade diagnosis for years and thus treatment options (1, 4)? In many cases, we’re simply not looking.
Confronted with severely ill patients presenting non-specific or overlapping symptoms, clinicians are burdened with choosing the optimal genetic test, that specific gene panel that will unearth an explanation. Unsurprisingly, with little to inform them, the narrow scope of gene panels rarely render an answer. This is especially true for copy number variation (CNV) associated pathologies. CNVs, the deletions or duplications of large chunks of DNA, are critical to genetic diversity and may even play a bigger role than single nucleotide polymorphisms (SNPs) in driving human adaptation and diversity (2). As such, many CNVs are benign. However, an increasing number are being associated with rare disease formation, both mendelian and possibly sporadic (2). While CNVs can be detected via multiplex ligation-dependent probe amplification (MLPA) and microarray, these laboratory assays can be costly and time-consuming. Thus, only a select subset of genes are often tested.

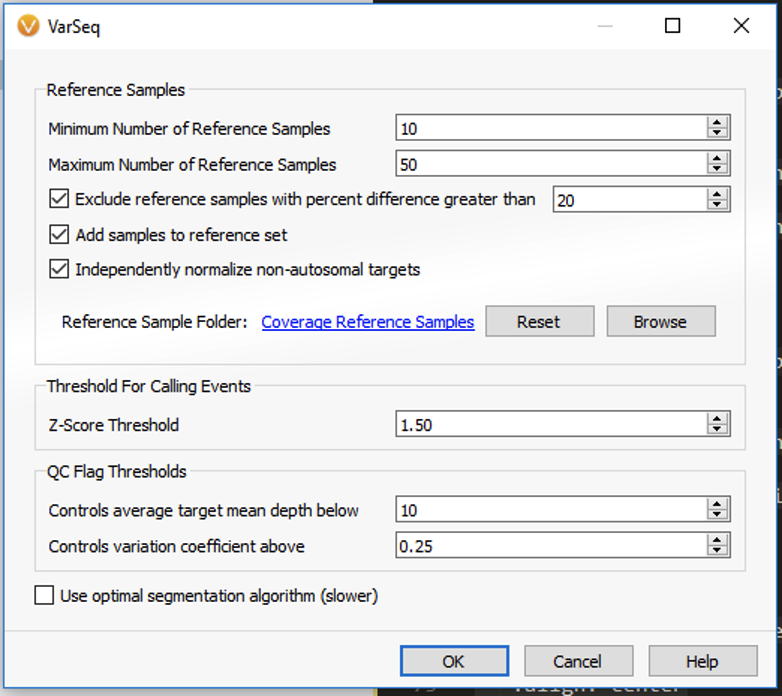
One solution is to eliminate the burden of choice. As NGS continues to become cheaper, faster, and more data-rich, whole-exome and especially whole-genome sequencing are becoming mainstream, even essential, for diagnosing rare diseases, 10% of which are estimated to be caused by CNVs (2). Challenges in CNV detection, such as short read length and GC bias, are being overcome through algorithms like VarSeq’s CNV Caller capable of detecting events of all sizes, unlike MLPA and microarray, based on metrics like ratio and z-score.
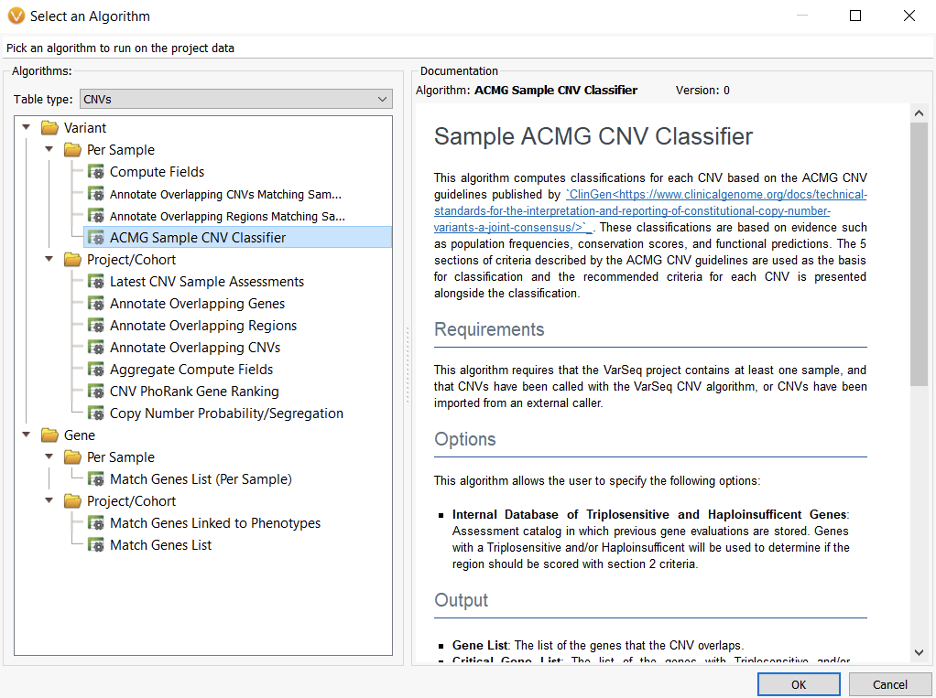
As we learn more about the variety of CNV molecular mechanisms, including gene dosage sensitivity, interruption or fusion at breakpoint junctions, regulatory element deletion, recessive allele exposure, and impact on noncoding elements like promoters and enhancers, we can no longer hope to find answers for the undiagnosed in just a handful of genes. Instead, we can radically increase diagnostic yields in less time by leveraging genomic scale data and automating the laborious classification process with VSClinical’s ACMG CNV classifier.
Clinicians can be equipped with the information they need for a diagnosis allowing kids with rare diseases the opportunity to read the Odyssey rather than live it. If you have any questions regarding diagnostic odyssey, please reach us at support@goldenhelix.com.
- Nguengang Wakap, S., Lambert, D.M., Olry, A. et al. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur J Hum Genet 28, 165–173 (2020).
- Research Topic: Copy Number Variation in Rare Disorders. Front. Genet. Editors: Bene, J., Komlosi, K., Gyenesei, A. (2021). https://www.frontiersin.org/research-topics/15268/copy-number-variation-in-rare-disorders#overview
- Dong, D., Chung, R.YN., Chan, R.H.W. et al. Why is misdiagnosis more likely among some people with rare diseases than others? Insights from a population-based cross-sectional study in China. Orphanet J Rare Dis 15, 307 (2020).
- Sung, H., Ferlay, J., Siegel, R.L., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J. Clin. 71, 209-249 (2021).