Today, we will be exploring the indispensable role of GenomeBrowse in your VarSeq workflows. This blog aims to guide you through various customization options, including color preferences, filtering techniques, display modifications, numeric value plotting, and additional tips. I hope to enhance your GenomeBrowse experience and enable you to gain valuable insights into your data. Let’s jump in by talking about… Read more »
Enhancing VarSeq Customization and Automation with Visual Studio Code: A Guide to Evaluation and Reporting Scripting Are you as excited as we are about the new automation and customization features we’ve been rolling out with VarSeq’s most recent release, VarSeq 2.3.0? Do your eyes light up at the prospect of automated integration of various data sources into VarSeq’s evaluation and… Read more »
In our previous blog, we covered the highlights of our Advanced Report Customization in VSClinical webcast in the context of germline clinical reports. Now, we bring you the next of the series: somatic clinical reports. In the recent webcast, Advanced Report Customization, we covered a range of somatic-focused clinical reports, demonstrating how easy it is to create AMP guideline-based clinical… Read more »
When it comes to clinical variant analysis for germline variants using ACMG guidelines, we understand that the clinical report is essentially the receipt for your services for the patient. In our recent webcast, Advanced Report Customization in VSClinical, we displayed several examples of report templates to show off the range of possibilities our users have to format their clinical report…. Read more »
As we move towards the end of the year, our FAS Team is excited to announce our short blog series highlighting some of the Customization Features in our up-and-coming VarSeq release! The goal of this blog series is to show examples of how generating a clinical report can be customized to accommodate a wide range of functionality. Our December webcast,… Read more »
The first two blogs in this series covered Sentieon’s somatic variant calling from Tumor with Normal (Part I) and Tumor without Normal (Part II). In addition to providing multiple somatic variant calling processes, users also have access to high-sensitivity scripts and full support for the GRCh38 reference assembly. Without going into excessive detail, this final blog of the series will… Read more »
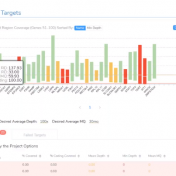
Our customers often ask if they can include quality control metrics in their final reports. While which metrics you actually need to report may be unique to your lab, there are a variety of metrics that we can immediately render into a report, and even more that can be rendered with a few customizations. For help with any report modifications,… Read more »
Get your year started off right with Golden Helix’s new VarSeq Onboarding and Training Program! Analysis of NGS data comes with several challenges, but mastery of the data analysis software does not have to be one of them. With this in mind, Golden Helix is pleased to announce that VarSeq Onboarding and Training tools are now available to all our… Read more »
Your variants of interest have been identified, the SVs annotated, and the CNVs classified. Once the manifest has been imported (and here is a great blog on the subject), the last step is to bring all of this information into a report template! In this blog, we will go over some simple report modifications and provide resources for more information… Read more »
…so we added some automation to your automation so you can automate while you automate! Automation has been a hot topic recently and for all the right reasons. As we (proudly) watch our customers increase their sample and data volume, we are constantly seeking to provide tools to reduce click rate and optimize throughput. Furthermore, with all of the new… Read more »
The ability to import patient-level information into VarSeq and VSClinical can not only save time while evaluating a sample but can be a critical step in automating project creation through VSPipeline. Here we will review how to construct a basic text manifest and how to apply that manifest to a project. Those fields can then be rendered into the final… Read more »
Our FAS team would like to thank everyone who attended our December 2022 webcast, A User’s Perspective: Somatic Variant Analysis in VarSeq 2.3.0. This webcast allowed three members of our FAS team to give their unique insights concerning the improvements to our new VarSeq 2.3.0 release, which will be recapped here. Starting with template creation, our Technical Field Application Scientist,… Read more »
Thank you to those who attended our webcast on the user perspective of our automated AMP guidelines! Furthermore, let me express our appreciation for those particularly engaged users who posed some very thoughtful questions. While we weren’t able to answer all of them live, I hope to shed some light on some pertinent details of somatic analysis here. Let’s start… Read more »
Oftentimes, the endpoint of a clinical variant analysis is a standardized, clinical report. As such, we ship a number of default templates with VSClinical for users to report their findings. But these templates are just a starting point! Our platform allows users to fully customize their reports to adhere to lab-specific preferences. We have shared a plethora of how-to’s on… Read more »
Although best known for its auto-generation of custom reports, VarSeq comes with a slew of options for exporting your data. In this blog, we will review some of the lesser-known methods for exporting your data into usable formats. These four export options can all be found under the Export tab at the upper left corner of your VarSeq interface (Figure… Read more »
Tumor profiling via next generation sequencing (NGS) often reveals secondary germline variants that may constitute important incidental findings. In May 2021, the American College of Medical Genetics and Genomics (ACMG) released an updated policy statement for reporting incidental findings in exome and genome sequencing data along with a corresponding list of genes. These recommendations state that laboratories should report pathogenic… Read more »
I’m sure everyone’s favorite winter activity is getting cozy next to a fire, with a nice cup of tea, and reading the VarSeq instruction manual, right? Of course all VarSeq users have read the manual, but if you have not had the chance to review this captivating read, let me entice all of you to do so by announcing that… Read more »
The recent release of VarSeq 2.2.2 brings our Word report template system, previously featured in VSClinical AMP, to the VSClinical ACMG workflow. This blog post will describe how to use the Word template system using one of our shipped templates as well as how to start customizing your own templates. We will cover the three different report templates that ship… Read more »
Webcast Recap In the recent webcast “Exploring New Features and Clinical Reports in the ACMG Guideline Workflow”, Gabe and I took viewers through an evaluation with CNVs and SNVs according to the ACMG Guidelines where we generated and customized a clinical report. Along the way, we highlighted many new features that will soon be available in the upcoming VarSeq release…. Read more »
In our previous webcast, Evaluating CNVs with VSClinical’s New ACMG Guidelines, we focused on a CNV deletion (12:27715515-29628122×1) in which the patient had a known disorder called Brachydactyly type E. The CNV was isolated using our VS-CNV caller and applied to the ACMG CNV guidelines using the intuitive steps of VSClinical. If you missed the webcast, you can watch the… Read more »