As many of our users know, VarSeq comes shipped with various project templates that are designed to give users a baseline workflow to get started with their projects. These templates are tailored for various applications including tumor-normal, trios, cancer and hereditary gene panels, and ACMG Guidelines workflows.
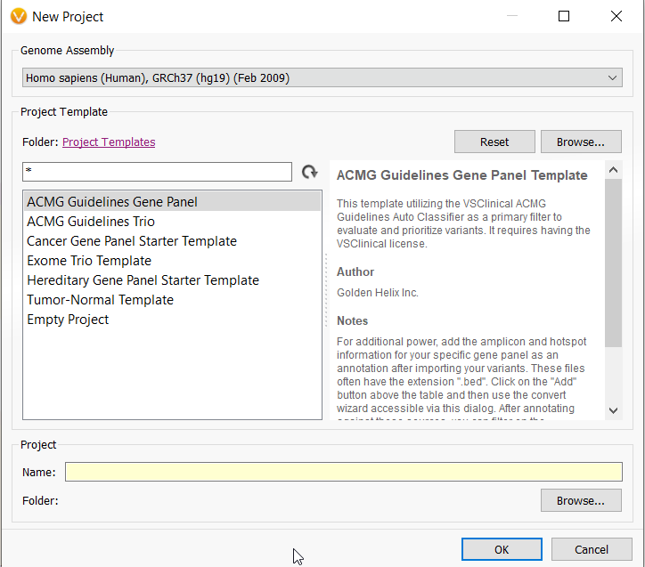
The templates contain application-specific annotation sources and algorithms that will automatically load into the project. Accompanying the databases and algorithms is a pre-built filtering logic constructed by leveraging fields from those databases. The filter chain will easily narrow the search to the most clinically relevant variants.
As mentioned, the purpose of the templates is to provide a basic workflow, and all users need to do is supply the data. However, as you begin to become more familiar with VarSeq over time, you may find that you want to include additional filters from other databases and algorithms… no problem! You can easily add, remove, or rearrange filters to customize your variant search. From there, you can save your project as a template to select the next time you start a new project and apply this workflow that you have customized.
For many readers, the value of using VarSeq project templates is well known. However, with more and more researchers and clinicians switching over to using the GRCh38 human reference genome, there was a need to have project templates designed for this reference assembly. The same project templates that were built using the GRCh37 reference genome are now available in GRCh38 format!
Perhaps you are thinking about switching over to using the GRCh38 reference genome, but you have data and custom annotations formatted with the GRCh37 reference genome assembly. Fortunately, Golden Helix has implemented a UCSC LiftOver algorithm in VarSeq so you can properly liftover your data and annotations.
If you are planning on switching, you can liftover GRCh37 data to GRCh38 in VarSeq by creating a new project and selecting the GRCh38 genome assembly in the dropdown at the top of the window.
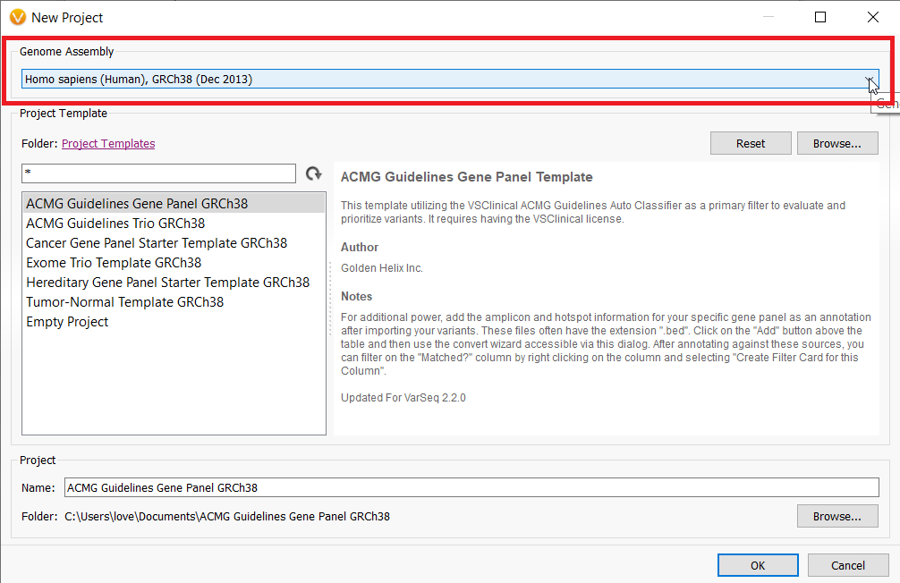
When you select GRCh38 as your genome assembly, the project templates become GRCh38-based so you can select your desired project template at this point as well.
Then follow the import wizard to import your data files. Once you get to the final step of the import wizard, be sure to check the “Liftover” box and enter GRCh37 for the assembly of input files.

Now, you are all set with GRCh38 project templates containing GRCh38 annotations and GRCh38 data! Our VP of Product & Engineering, Gabe Rudy, discusses the costs and benefits of the GRCh38 reference genome and the Liftover tool in more detail in this webcast and blog for anyone interested in learning more. Alternatively, if you would like to talk to our team directly, please contact us at support@goldenhelix.com!