In the rapidly evolving field of genomics, innovative technologies, and methodologies are constantly being developed to enhance our understanding and diagnosis of genetic disorders. Golden Helix’s VarSeq and VS-CNV software platforms stand at the forefront of this revolution, offering powerful tools for genetic data analysis and interpretation. This blog highlights recent research endeavors where these tools have been instrumental. From… Read more »
In our recent webcast, we unveiled the integration of the Twist Bioscience Exome 2.0 Plus Comprehensive Exome Spike-in capture panel with VS-CNV, marking a significant advancement in genetic diagnostics. By addressing the limitations of standard exome kits that miss vast genomic regions, our enhanced panel introduces ‘backbone’ probes for comprehensive genomic coverage. This innovation enables the detection of CNVs, LOH,… Read more »
Every month we compile customer publications that reference us, and every time I am excited to see the amazing work being done around the world. From pediatric heart conditions to rare diseases, or a Thai clinical study on dilated cardiomyopathy, it is always a pleasure to see the Golden Helix Software suite helping research and clinical genetics. Below are a… Read more »
Below is a list of highlighted peer-reviewed publications from this month. The Golden Helix team consistently enjoys seeing our software applications hard at work in the field, whether it is clinical whole-exome sequencing, targeted CNV identification, or genotype-phenotype correlations. Enjoy reading below how our suite of VarSeq, VSClinical, and VS-CNV continues to contribute to the progression of genetics research. Looking… Read more »
If you stay current on the developments of Golden Helix features, you are aware of the substantial evolution of our copy number detection and evaluation capabilities in VarSeq. The process of CNV detection and evaluation is typically handled through the VarSeq graphic user interface. However, in some cases, users benefit from running this process via the command-line interface. Fortunately, Golden… Read more »
The VarSeq CNV calling algorithm, VS-CNV, is a powerful tool for calling CNVs from the NGS coverage data stored in your BAM files. However, before this algorithm can be deployed in a clinical setting, it must be tuned and validated using data that is representative of your lab’s NGS workflow. In the past, this validation process could be difficult, as… Read more »
Established in 2004 and headquartered in Chennai, India, with regional centers across the country, LifeCell runs India’s largest stem cell bank and has also diversified into diagnostics and tissue therapeutics. They employ roughly 2,000 people, providing genetic services to customers in the mother and baby space. Phani Nagaraja Setty is working as a scientist at LifeCell. Setty obtained his Master’s… Read more »
As the world is consumed by the ongoing pandemic, it is easy to forget that there are investigators all around the globe that continue to make important discoveries in human medicine. Below are a few examples that remind us there are those that persevere in their chosen fields of study despite the trying times. At Golden Helix, we continue to… Read more »
The detection and interpretation of Copy Number Variants (CNVs) is vital for the clinical evaluation of individuals with a wide range of disorders. Golden Helix has remained at the forefront of CNVs in Next-Gen Sequencing (NGS) data since 2016 with the release of VS-CNV, our solution that allows you to both detect and analyze CNVs directly from NGS data. Earlier… Read more »
We have had many customers come to us over the years with a simple problem: they have BAM files for whole exome or gene panel data and would like to call CNVs using VarSeq’s powerful CNV calling capabilities, but they don’t have a bed file defining the target regions for their samples. To address this problem, we have developed a… Read more »
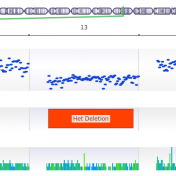
Copy Number Variation (CNV) is a type of structural variation in which sections of the genome are duplicated or deleted. Although CNV events are rare in the human population, constituting approximately 10% of the human genome, they are also associated with being causal mutations for disease phenotypes. Because of this, it is important for clinical and research settings to identify… Read more »
The common approaches to detecting copy number variants (CNVs) are chromosomal microarray and MLPA. However, both options increase analysis time, per sample costs, and are limited to the size of CNV events that can be detected. VarSeq’s CNV caller, on the other hand, allows users to detect CNVs from the coverage profile stored in the BAM file, which allows you… Read more »
2017 was a busy year regarding the development of our CNV tools. Since the release of the CNV caller, we have produced quite a bit of content tailored to assist our users with getting started. Here are some links: Robarts Research Institute CNV analysis on patients with familial hypercholesterolemia CNV annotations Common CNV questions CNV calling with shallow whole genome… Read more »
First-place Abstract Competition Winner, Michael Iacocca, shared his research with the Golden Helix Community during our February webcast ‘Using NGS to detect CNVs in familial hypercholesterolemia‘. In this webcast, he gave a great explanation on how our CNV caller aided his team in their research. If you were unable to join us for the event, you can find a recording… Read more »
One of our main focuses in 2017 was VS-CNV which allows clinicians to directly call CNVs in target regions quicker, easier and more affordably than CMA or MLPA testing. Our clients at Robarts Research Institute shared their recent publication with me which confirms that our time and dedication to our CNV capabilities was well worth it. I am delighted to… Read more »
This month we hosted two, incredible webcasts officially announcing the latest CNV annotation capabilities our Software Engineering Team has been hard at work on for the past couple of months. Our first webcast, Comprehensive Clinical Workflows for Copy Number Variants in VarSeq, was presented by Golden Helix’s VP of Product & Engineering, Gabe Rudy, who reviewed the expanded capabilities of… Read more »
Gabe Rudy gave a fantastic presentation yesterday on the latest additions to VS-CNV annotations. If you weren’t able to join us for the live event, you can access the recording and webcast slides here: Comprehensive Clinical Workflows for Copy Number Variants in VarSeq. Additionally, there were many great questions asked that we wanted to share with the community. Question: Can I… Read more »