Tag Archives: SVS
Third-Place Abstract Competition Winner

I would like to congratulate our third and final winner of the 2017-18 Abstract Competition, Suxu Tan, a current PhD Student at Auburn University. Our first and second place winners, Michael Iacocca and Nicole Weaver, performed their research using VarSeq. Suxu, however, used SVS to conduct the research in his paper ‘GWAS analysis of QTL for resistance against Edwardsiella ictaluri in… Read more »
Genomic Prediction Methods in SVS Q&A

Yesterday’s webcast, Genomic Prediction Methods in SVS, gave attendees a chance to see how the principles of genomic prediction are applied within SVS, predicting phenotypes for both plant and animal species. You can find a recording of the webcast on our site here should you be interested in checking it out or sharing with a colleague! The webcast garnered a… Read more »
Customer Publications From Around the Globe

It’s fascinating to hear the various ways our products are being used by customers all around the world. This month we’re featuring publications citing VarSeq, SVS and HelixTree which cover studies from Schizophrenia, Multiple Sclerosis and more. We hope you enjoy! Exome sequencing in schizophrenic patients with high levels of homozygosity identifies novel and extremely rare mutations in the GABA/glutamatergic pathways This… Read more »
GWAS Workflows with SVS webcast Q & A

In case you missed our live event yesterday, I wanted to share a link to the webcast recording: New Enhancements: GWAS Workflows with SVS. There were several questions asked, so we’ve also shared the Q & A session below! Question: Are these enhancements priced as a separate feature? Answer: No, SVS is a constantly evolving platform, so everything you see in this webcast is… Read more »
Webcast: New Enhancements – GWAS Workflows With SVS

We are dedicating our August webcast to explain the latest and greatest enhancements with GWAS workflows in SVS. Below you can find further details and registration information. We hope you can attend! Wednesday, August 9th, 2017 12:00 PM EST In this webcast, we will focus on the recent improvements to our research product SNP & Variation Suite. Over the past… Read more »
Understanding Your GWAS Signal with LD Scores
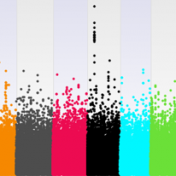
When studying complex diseases that may have multi-genic contributions from across the genome, it is not uncommon to see what may appear like elevated correlation between your trait or other test variable and the SNPs across the genome. The problem is at first glance you won’t be able to tell if this is due to a population structure in your… Read more »
Recent Customer Success

As usual, we would like to share some of the articles that our clients have published recently citing our software. These articles are incredibly diverse and interesting, and we hope you enjoy browsing through them. Fielding Hejtmancik and Lin Li of the NIH-NEI and colleagues published Homozygosity Mapping and Genetic Analysis of Autosomal Recessive Retinal Dystrophies in 144 Consanguineous Pakistani… Read more »
Recent Publications from SVS Users

We had lots of customers publish their work using our SVS software, and I wanted to share their work with you. Congrats to all! Here are some of the highlights: Francesca Fernandez of the University of Wollongong along with colleagues published Effects of common GRM5 genetic variants on cognition, hippocampal volume and mGluR5 protein levels in schizophrenia in Brain Imaging and… Read more »
PhoRank in SVS: Gene Ranking for Your Research Genotypes

Since we released our Phenotype Gene Ranking algorithm in VarSeq, it has become a staple of the way people conduct their analysis. It allows for a combination of filtering with ranking to prioritize follow-up interpretations of analysis results. Our PhoRank algorithm will be available in our upcoming SVS release to also aid in the numerous research workflows performed on SNPs… Read more »
Massive Variant Boost to ClinVar & PubMed Citation Fields

It may have been easy to miss in the drum-beat of monthly annotation updates we do here at Golden Helix, but there are a couple of things that are very special about the January update to the ClinVar database: We added new fields including HGVS names of variants and citations in PubMed for variants ClinVar nearly doubled in size by… Read more »
Determining the best LD Pruning options
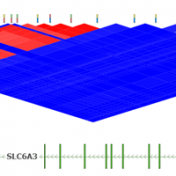
Pruning your data based on Linkage Disequilibrium (LD) values is an important quality assurance step for GWAS analysis. In particular, some tests such as Identity by Descent Estimation (IBD), Inbreeding Coefficient Estimation (f) and Principal Component Analysis (PCA) will obtain better results if the markers used are not in linkage disequilibrium with each other. Therefore, Golden Helix’s SVS provides the… Read more »
Compute Kinship Matrices & GBLUP on Very Large Sample Sets

Now available in SVS! Increasingly important in the analysis of the genotype to phenotype relationship is accurately accounting for the relatedness of samples. This is especially important to model correctly in plant and animal populations where man-directed breeding shapes the relationship structure. Along with trait association, one of the high-value use cases for genotyping animals and plants is to estimate… Read more »
Understanding the genetic mechanisms of inherited eye disease at the NIH – NEI

Dr. James Fielding Hejtmancik and his team at the National Eye Institute’s Ophthalmic Genetics and Visual Function branch, use linkage and association studies to better understand the genetic mechanisms of inherited eye disease. The team investigates both Mendelian and complex diseases as well as functional work with a focus on cataracts, retinal degenerations, myopia and corneal dystrophies. “If it affects… Read more »
Guest Presentation by Dr. Folefac Aminkeng

Dr. Folefac Aminkeng, first place winner in this year’s Abstract Challenge, will present his work to the community via a live webcast on Wednesday, April 20th at noon EDT. Dr. Aminkeng is a Postdoctoral Fellow at The Centre for Molecular Medicine and Therapeutics (CMMT) at the University of British Columbia in Vancouver, BC. Aminkeng’s research focus is the pharmacogenomics of adverse… Read more »
The Golden Helix Soccer Team

The Golden Helix Soccer Team I recently received a very special email from a long time Golden Helix customer, Dr. Vanessa Hayes of the Garvan Institute of Medical Research in Sydney, Australia. In 2010 Dr. Hayes, then of the J. Craig Venter Institute, joined several colleagues in collaboration on The Southern African Genome Project. The project was aimed at including African genomes in existing… Read more »
New SVS Meta-Analysis Example Project
New SVS Meta-Analysis Example Project The latest version of the GWAS E-book featured a chapter on Meta-Analysis. We are pleased to make the project used in this chapter available as an example project for SVS (SNP & Variation Suite). This blog post will walk you through the analysis steps used in the example project. This information is also contained in… Read more »
GWAS Example Project Updated for SVS Viewer
With the release of our updated GWAS E-book, we have recently updated the GWAS example project (SNP Genome-Wide Association Tutorial – Complete). This updated project includes more details about how spreadsheets were generated, how to generate plots and which images were used for the GWAS E-book. This information can be found in the User Notes view in the project navigator and… Read more »
What to expect from Golden Helix in 2016

Golden Helix in 2016 We had a terrific year 2015. It was the year in which we got serious about the clinical testing market. We successfully continued on the path of attracting more referenceable clients such as University of Iowa, Baby Genes, Prevention Genetics and many more. We rounded out our VarSeq suite by adding more clinically relevant features and… Read more »
Visualizing Meta-Analysis results with a Forest Plot

We have just released SVS version 8.4.2, and included in the release is a new script for visualizing Meta-Analysis results with a Forest Plot. You can find full details on all the new and updated features included with the update in our Release Notes. Release Notes from all our software products can be found on our Support Bulletin web page… Read more »
