I remember visiting a patient in the NICU amongst the incubators, some glowing blue like tiny tanning beds to treat jaundice, all containing tiny humans – many smaller than a loaf of bread. Infants get admitted into the neonatal intensive care unit or NICU for many reasons ranging from elevated bilirubin, hypoglycemia, sepsis, and respiratory distress (RDS). Many are eventually… Read more »
Thank you to those who attended our recent webcast, “PhoRank 2.0: Improved Phenotype-Based Gene Ranking in VarSeq”. For those who could not attend, you can find a link to the recording here. This webcast covered upcoming improvements to the PhoRank phenotype-based gene ranking algorithm based on literature published in the years since the algorithm’s development. The PhoRank Algorithm When performing… Read more »
The PhoRank tool in VarSeq is further explored in this post by looking at the sample-specific capability. VarSeq PhoRank Part: 1 Variant Phorank Gene Ranking showed how the PhoRank algorithm could be applied to all the variants in a VarSeq project, regardless of the number of (or difference in) samples. There is another PhoRank algorithm in VarSeq that allows the… Read more »
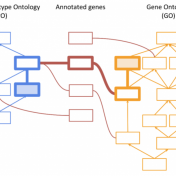
One of the main goals of clinical genomic labs is to identify problematic variants in affected individuals. One tool to assist in this search is the phenotype driven variant ontological re-ranking tool in VarSeq called PhoRank. A common situation facing clinicians is sorting through thousands of variants provided by an individual’s exome data (or possibly the individual’s nuclear family exome… Read more »
Recently, we added a natively supported Genotype Phasing and Imputation capability in SNP & Variation Suite 8.7.0. Since then we have had fantastic feedback and adoption as folks take advantage of the BEAGLE 4.0 and 4.1 algorithms from within their existing SNP GWAS and agrigenomic workflows. One piece of feedback we heard from our time at PAG, ACMG and our… Read more »
Since we released our Phenotype Gene Ranking algorithm in VarSeq, it has become a staple of the way people conduct their analysis. It allows for a combination of filtering with ranking to prioritize follow-up interpretations of analysis results. Our PhoRank algorithm will be available in our upcoming SVS release to also aid in the numerous research workflows performed on SNPs… Read more »
I am constantly on the lookout for fun or interesting datasets to analyze in SVS or VarSeq and recently came across a study looking into inherited cardiac conduction disease in an extended family (Lai et al. 2013). The researchers sequenced the exomes from five family members including three affected siblings and their unaffected mother and an unaffected child of one… Read more »
Personal genome sequencing is rapidly changing the landscape of clinical genetics. With this development also comes a new set of challenges. For example, every sequenced exome presents the clinical geneticist with thousands of variants. The job at hand is to find out which one might be responsible for the person’s illness. In order to reduce the search space, clinicians use various methods… Read more »