Recently, we added a natively supported Genotype Phasing and Imputation capability in SNP & Variation Suite 8.7.0. Since then we have had fantastic feedback and adoption as folks take advantage of the BEAGLE 4.0 and 4.1 algorithms from within their existing SNP GWAS and agrigenomic workflows. One piece of feedback we heard from our time at PAG, ACMG and our… Read more »
Our webcast series for 2017 is starting off by giving the Golden Helix community their first look at our addition of genotype imputation into SVS! On Wednesday, January 11th, Gabe Rudy will discuss how users can now run their genotype phasing and imputation on human and animal data as part of their SVS analytics workflow. Wednesday, January 11th @ 12:00 PM, EST Genotype imputation… Read more »
Genotype imputation is a statistical technique for estimating sample genotypes at loci that were not directly assayed by sequencing or microarray experiments. There are several reasons why you might want to use imputation in a research study. For example: Improve call rates in GWAS by imputing sporadic missing genotypes Harmonize the data content from different GWAS genotyping platforms so that… Read more »
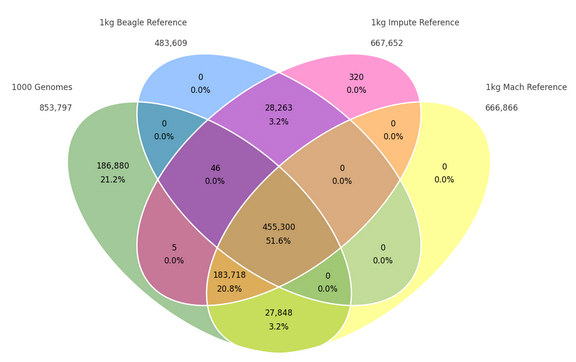
Genotype imputation is a common and useful practice that allows GWAS researchers to analyze untyped SNPs without the cost of genotyping millions of additional SNPs. In the Services Department at Golden Helix, we often perform imputation on client data, and we have our own software preferences for a variety of reasons. However, other imputation software packages have their own advantages… Read more »