If you stay current on the developments of Golden Helix features, you are aware of the substantial evolution of our copy number detection and evaluation capabilities in VarSeq. The process of CNV detection and evaluation is typically handled through the VarSeq graphic user interface. However, in some cases, users benefit from running this process via the command-line interface. Fortunately, Golden… Read more »
In our previous webcast, Evaluating CNVs with VSClinical’s New ACMG Guidelines, we focused on a CNV deletion (12:27715515-29628122×1) in which the patient had a known disorder called Brachydactyly type E. The CNV was isolated using our VS-CNV caller and applied to the ACMG CNV guidelines using the intuitive steps of VSClinical. If you missed the webcast, you can watch the… Read more »
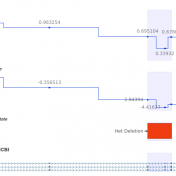
In trio workflows, one of the most important factors in scoring a variant is understanding how that variant is inherited from the parents. Likewise, when looking at extended families, the segregation, or presence of the variant among the affected versus unaffected individuals provides evidence for its pathogenicity for a given phenotype or disease. Given the nature of Copy Number Variants… Read more »
We are happy to announce that our latest version of SVS includes the ability to call CNVs on low read depth Whole Genome Sequencing (WGS) data. Designed for calling large cytogenetic events, this algorithm can detect chromosomal aneuploidy events and other large events spanning one or more bands of a chromosome from genomes with average coverage as low as 0.05x…. Read more »
We just got back from three busy days at the Molecular Pathology (AMP) conference in friendly San Antonio, Texas. Keeping up the Golden Helix conference momentum for the year, we had 3-4 in-booth demonstrations a day covering our CNV calling, variant interpretation, and data warehousing products for NGS-based genetic tests. And in short, NGS based tests for cancer and germline… Read more »
Our abstract competition is one of my favorite events because of the learning opportunity it provides our team. Each applicant’s submission tells us a unique story about how our software is helping users conduct their research. This year’s competition didn’t disappoint bringing a new round of fascinating studies to our attention. However, with all these great abstracts came the difficult… Read more »
Low read depth? Great! We are excited to introduce our new CNV calling algorithm for low and ultra-low read depth Whole Genome Sequencing (WGS) data. This algorithm is designed to call large cytogenetic events with high confidence from low read depth whole genome data, with as few as one million aligned reads or 0.02x coverage. The low sequencing cost of… Read more »