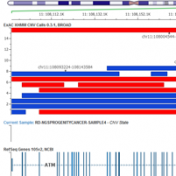
ExAC CNVs were released publicly with a recent publication, providing the full set of rare CNVs called on ~60K human exomes. While there are many public CNV databases out there, this is the first one that was derived from exome data, and thus includes both extremely rare and very small CNV events. With the recent release of Golden Helix’s CNV calling… Read more »
Dr. Laura Li and her colleagues at the Children’s Hospital Los Angeles (CHLA) are working to determine the underlying genetic causes of Optic Nerve Hypoplasia (ONH), which is still unclear. ONH is the absence or under-development of the optic nerve and is currently the leading ocular cause of vision impairments and blindness in young children. ONH can also be combined… Read more »
We hope our Golden Helix community had a delicious Thanksgiving, and several of our customers had an additional reason to celebrate because they published this month, including one book publication! The success is overwhelming and we are happy to share it with you each month: Randovan Kasarda of the Slovak University of Agriculture and colleagues published Genetic Divergence of Cattle Populations… Read more »
December’s webcast will provide the Golden Helix community with a more in-depth look at CNV analysis in VarSeq. On December 7th, Dr. Nathan Fortier will discuss the challenges and metrics surrounding CNV detection and then demonstrate VarSeq’s new capability from VCF to clinical report. Wednesday, December 7th @ 12:00 PM, EST Numerous studies have documented the role of Copy Number Variations (CNVs)… Read more »
True to its nature, VarSeq offers multiple data export options. You can export result tables from VarSeq to Text, VCF, a VarSeq annotation file and most importantly an XLSX (Excel) File. VarSeq’s Excel export options provide a lot of flexibility in the information that is exported and preserve the formatting of data during the export process from VarSeq to Excel. This… Read more »
While clinical assessments of germline mutations have been collected in ClinVar under the stewardship of the NCBI and the collaborate effort of many testing labs, the same type of resource has been missing for mutations that could informal clinical care in Cancer. Or at least, that is what I thought until I started to work with CIViC. With the stewardship of… Read more »
This year we have added some very important features to our software including CNV calling in VarSeq and integrating our premium annotations into SVS. We have also made many improvements to our software’s performance so that you can handle growing datasets with relative ease. In an effort to thank our community for their support throughout the year, we are happy… Read more »
One of the tools at the top of the toolbox for researchers working with microarray data is genotype imputation. Genotype imputation is the process of inferring the genotype of one or more markers based on the correlation pattern (aka linkage disequilibrium or LD) of the surrounding markers for which genotypes are known. We have now integrated a natively ported version of BEAGLE into Golden… Read more »
We closed October with a record number of publications from our customers! Topics ranged widely from root traits in tropical maize to pretreatment depression severity. We hope you’ll enjoy reviewing some of our favorites: Bradley Aouizerat of NYU and colleagues published Human leucocyte antigen class I and II imputation in a multiracial population in the International Journal of Immunogenetics which considered… Read more »
This month’s webcast, Agrigenomics 2.0 – Advanced Analysis to Accelerate Discovery, will feature two well known Agrigenomic researchers and long-time Golden Helix customers, Christopher Seabury of Texas A&M and Holly Neiburgs of Washington State University. These two will join our own Gabe Rudy for a look at advanced workflows in SVS to advance mammalian genetic research. We hope you can join us! Wednesday,… Read more »
ASHG 2016 is in our rear mirror. Again, it was bigger and better than the previous year. The conference hosted over 9,000 visitors from 66 countries. This gave the event a level of vibrancy that was evenly matched by the wonderful ambiance of the city of Vancouver. Nestled in between the two conference centers was a little pier offering spectacular… Read more »
The new Annotate and Filter algorithm is now available with the release of SVS 8.6.0, see the release notes for full details on all new and updated features. To access this new functionality, you simply need to update your SVS installation to the new version. The update can be done by clicking the Update Available link at the bottom of… Read more »
Dr. Sergey Kornilov, a Duncan Scholar in Molecular and Human Genetics at Baylor College of Medicine, combines his broad psychology background with genetics to research the genetic basis of neurodevelopmental disorders with a unique dual perspective. Neuro-developmental disorders, for example, those of the spoken and written language, affect many worldwide – up to 10% of preschool children. In most cases, these… Read more »
It’s hard to believe that summer has already faded into fall and that ASHG 2016 is right around the corner! 2016 has been quite a busy year for us so far at Golden Helix. We have been working hard to bring our community the very best tools available for variant interpretation and SNP analysis. This year at ASHG, you can… Read more »
Copy Number Variants have been important to clinical genetics for quite a while now. So, what has made now the right time to be looking at calling CNVs from NGS data? Well, there are a number of good reasons. The dominant one is simply that the NGS data you are already creating for calling variants can be used in many cases… Read more »
In our SVS 8.6.0 release, we updated our Annotate and Filter Variants feature to utilize our powerful VarSeq annotations. Annotations can be run against gene, interval, variant, and tabular tracks, including RefSeq, ClinVar, CADD, OMIM and OncoMD. The new streamlined dialog allows users to select track specific options and to set up custom filters. While our public annotation repository has… Read more »
We are excited to announce that our CEO & President, Dr. Andreas Scherer, was featured in “The 50 Most Creative CEOs to Watch” issue of Insights Success magazine this month. The article is focused on how Dr. Scherer is leading Golden Helix to deliver top bioinformatics solutions to further enable Precision Medicine. Please take a look at the article here.
Many of our customers published throughout September using our SVS software, and we love sharing their work with you. There’s bound to be a topic among the wide variety we’ve highlighted here that will spark your interest! Maurico Arcos Burgos, Claudio Mastronardi and colleagues of the Australian National University published Mutations modifying sporadic Alzheimer’s disease age of onset in the American Journal of Medical… Read more »
Copy Number Variations (CNVs) play an important role in human health and disease, and the detection of CNVs in clinical samples has the potential to improve clinical diagnoses and inform treatment decisions. Yet until now, if you wanted to have CNVs on your targeted gene samples, you would need an alternative assay such as Chromosomal Microarrays (CMAs). In this webcast,… Read more »
Gabe Rudy’s webcast yesterday, Big Data at Golden Helix: Scaling to Meet the Demand of Clinical and Research Genomics, was a huge success with well over 300 registered. In today’s blog post, I wanted to recap the Q&A session with Gabe. If you missed the webcast, check it out! Question: What is the end goal for an application like Warehouse? Answer: The… Read more »