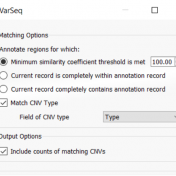
Thank you for attending the webinar focused on implementing VarSeq and VSClinical for family-based workflows. If you would like to use the webinar as a reference or were not able to attend, you can access it using the following link to view ‘Family-Based Workflows in VarSeq and VSClinical. Here is a brief recap of what we discussed: This webinar demonstrated… Read more »
VarSeq recently received major upgrades in a wide range of areas, one of these areas includes adding annotations such as GnomAD. This includes new fundamental methods of CNV ACMG guideline processing but also a large number of small additions in annotations. One addition is the application of gnomADs – Gene Constraints. This provides various metrics for pathogenicity on a per… Read more »
There is a multitude of interesting new features that have been incorporated into VarSeq 2.2.2. In this blog, I want to continue the discussion of these features and how each can be incorporated into your workflow, and also discuss the application of the Probability Segregation algorithm for copy number variation (CNV) analysis. The Probability Segregation algorithm is a new algorithm… Read more »
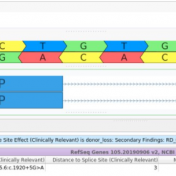
Our latest VarSeq release is one of the largest we’ve ever had, boasting an extensive list of new features and improvements. As part of this release, we have dramatically expanded our support for splice site analysis. This includes improvements to our novel splice site algorithm and support for splice site effect prediction along with several other small improvements. Novel Splice… Read more »
New discoveries using NGS data analysis are never-ending and are pushing precision medicine to the forefront. In this month’s customer publication blog, I am focusing on our VarSeq software as investigators harness its power to perform a variety of investigational study designs. From cancer to inherited rare disease research, VarSeq is the rising star in research and diagnostic tools. At… Read more »
I am pleased to share with you the official release of my updated eBook, “Precision Medicine“. Almost 2,500 years ago, Hippocrates captured one of the key principles underlying precision medicine. In the 21st century, we take an understanding of the individual characteristics of a person to a new level. By leveraging information about an individual’s genome, we are able to… Read more »
In continuation of our blog posts focusing on new features of VarSeq v2.2.2, here we will discuss the Latest Sample Assessment algorithm for both single nucleotide variants (SNVs) and copy number variants (CNVS). This algorithm annotates the variants of the project with the latest assessment from your variant catalog, which will show the history of interpretations made for the variants… Read more »
Annotating genomic variants is a very complex process but perhaps the most important part of next-generation sequencing variant analysis. Here at Golden Helix, we recognize the importance and value of having the most up-to-date sources available and curating new annotation sources as they become available for variant analysis. Golden Helix has curated over 100 annotation sources for human variant analysis… Read more »
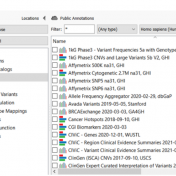
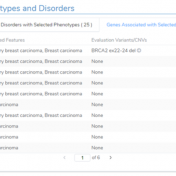
Our recent release of VarSeq 2.2.2 comes with a long list of upgrades and new features. In this blog post, we will demonstrate how defining sample phenotypes are available in VSClinical. One noticeable change is the ACMG guideline variant evaluation in VSClinical. Not only has this interface added CNV guideline evaluation, simplified the reporting process with embedded Microsoft Word and… Read more »
Didn’t catch the webcast live? No worries! We cover ‘VSClinical: A Complete Clinical Workflow Solution’ Q&A’s in this blog post. The webcast, ‘VSClinical: A Complete Clinical Workflow Solution’ demonstrated how solutions provided by Golden Helix can be implemented to cover all requirements of a clinical workspace. Specifically, this webcast focused on a detailed workflow from a bioinformatician, geneticist, and lab… Read more »
Golden Helix continues to innovate and grow, and due to this, we have been garnering attention and praise. We are thrilled to share with you some of the accolades and interest from numerous publications from the last six months. Talking points ranged from diagnosing and tracking Covid-19 infections leveraging NGS to new workflows, interpreting and reporting on copy number variants… Read more »
Golden Helix offers a market-leading bioinformatics solution that allows users to evaluate next-generation sequencing variants according to the ACMG and now ACGS guidelines. The ACMG guidelines were created in 2015 and are widely accepted as best practice for the interpretation of sequencing variants throughout the United States (Richard et al 2015). Very similar are the ACGS guidelines that were developed… Read more »
Happy New Year! I hope you had an opportunity to relax over the holidays with your family and friends. It’s time to start talking about our plans for the New Year, but first, please allow me to review a few highlights from 2020 that helped build a foundation for our future. Growth: Golden Helix was named to the 2020 Inc… Read more »
Have you seen us in The Journal of Precision Medicine? Last week, our team released VarSeq 2.2.2 loaded with a number of updates and VSClinical’s highly-anticipated ACMG-CNV Guideline workflow! We have spent the past several months sharing webcasts and blogs on this new capability. We are honored to also have this new solution recognized in The Journal of Precision Medicine… Read more »
VarSeq 2.2.2 was released on December 17th, 2020 and the main feature that was added to VarSeq was that the VSClinical ACMG Guidelines workflow now has an additional CNV interpretation framework based on the ACMG/ClinGen guidelines. This product supports interpreting CNVs detected with VS-CNV or imported CNVs alongside variants and requires both a VSClinical ACMG license and a CNV license…. Read more »
Reading scientific articles that our customers have recently published is one of my favorite things here at Golden Helix. It is fascinating to learn about the research and to see the various ways our software gets put to the test.Since we are rolling out our most extensive VarSeq update yet, I thought it would be a great time to look… Read more »
Our upcoming release of VarSeq is one of the largest we’ve ever had with our software! It comes with an extensive list of polishes and new features like our recently mentioned ACMG CNV classifier and a redesigned reporting interface with updated templates. Additionally, this new release is also paired with some major upgrades to our list of new and supported… Read more »
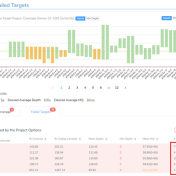
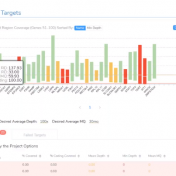
Curious about how coverage statistics can be used in conjunction with VarSeq? Evaluating the coverage over target regions or whole genomes is essential whether you are working with variant or CNV analysis. VarSeq has had the capability to compute sample level coverage statistics for some time now, but in the 2.2.2 release of VarSeq, there are some new features that… Read more »
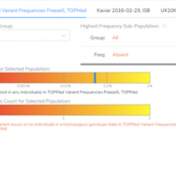
Our previous webcast demonstrated some of the new functionalities of VSClinical, including the ability to add ACMG frequency sources for the ACMG BA1, BS1, and PM2 criteria. This new feature was spurred by the feedback from our users, which requested supporting frequency tracks other than gnomAD Exomes and 1kG Phase3. Now, users can implement population catalogs to VSClinical such as… Read more »
Webcast Recap In the recent webcast “Exploring New Features and Clinical Reports in the ACMG Guideline Workflow”, Gabe and I took viewers through an evaluation with CNVs and SNVs according to the ACMG Guidelines where we generated and customized a clinical report. Along the way, we highlighted many new features that will soon be available in the upcoming VarSeq release…. Read more »