This week we launched VSClincial with our first webcast to show our powerful new way to perform variant interpretation following the ACMG guidelines. Our audience asked a lot of great questions on the new product and I’d like to highlight a few here.
Can VSClinical run on a laptop and/or a locked down environment?
Like all of Golden Helix products, you have complete control over how this is deployed. This doesn’t have to be deployed in a cloud environment and you are not uploading your data to us. That’s part of core business model and it allows us to set you up yearly license subscription and not monitor your usage, or track you on a per-sample basis. VSClinical is a part of VarSeq and deployed in that same model with the ability to run entirely offline. You would need to download some required annotation sources to run in an offline mode, or you can read some of those on-demand if you want to get up and going more quickly and are in an online environment.
Many genes have traditional or processed nomenclature, is there a way to set that nomenclature for each gene?
There are several things we’re bringing in on a per-gene basis. This includes links to OMIM, UnitProt, InterPro, Entrez and also includes a gene summary and a list of previously used gene symbols. You saw during the demonstration that we were detected commonly annotated phenotypes that are being associated with each gene. If that specific customization isn’t in there, keep in mind that this is our initial release and we’re working with our customers to make it customized to their needs, so we’d be happy to talk to anyone who would like to see a specific customization implemented.

Screenshot of the Gene and Transcript section of VSClinical, demonstrating many per-gene resources that are referenced by the software.
Will some of these new algorithms such as splice-site predictions be available outside of VSClinical?
Splice-site prediction will be able to run not just inside of that interactive interpretation but also as an algorithm that annotates all the variants in your project. It will require one of the VSClinical packages, but there are several options in terms of licensing structure, so just talk to us about your needs and we can make sure to make that work even if you’re not using all of VSClinical’s capabilities.
What is the typical analysis time for a sample where the phenotype includes something more complex such as ASD or another neurodevelopmental disorder?
That is a really great question, and this is obviously going to be a lab specific answer. I can’t answer to what the average times are, but I think at a high-level, one thing I think this eludes to is that it is important for labs to have introspection into how long things take: how long does it take on average for us to analyze a variant in this gene panel vs this other one, where are we spending our time, etc.
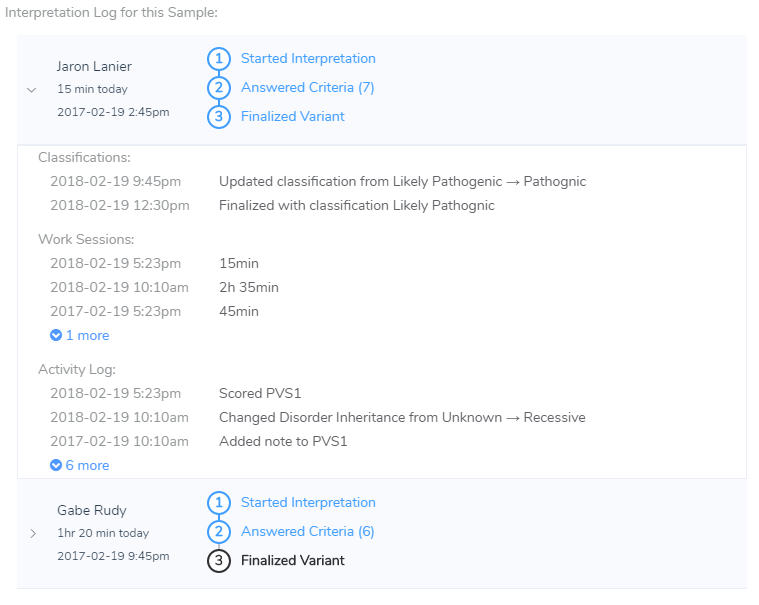
Example interpretation log for a given sample. The individual activities are tracked, but also the important milestones of classifying a variant and even how much time is spent in each work session on a given variant.
So one thing VSClinical is going to strive for is not only being CLIA compliant in the sense that we will log who is performing all of the actions of the analysis, but we will also want to support tracking how much time is spent evaluating variants and eventually allow you to look at these trends in the aggregate so you know what types of variants or situations end up dominating your analysis time.
If you have any questions on VSClinical or would like to see a personalized demonstration of the product to see how it works, get a hold of us by emailing info@goldenhelix.com!