Yesterday we launched VSClincial with our first webcast in what will be a series about this powerful new way to perform variant interpretation following the ACMG guidelines. In this post, I wanted to cover the motivation for VSClinical and how we curated and presented the 33 criteria from the ACMG Guidelines into an intuitive workflow with various bioinformatic evidence and supporting literature references to allow experts and starting variant scientists to tackle the most daunting of variants.
Variant Interpretation – the Keystone of a Genetic Test
There are currently 26K NGS based genetic tests registered on the NIH Gene Test Registry. One thing they all share is the potential to detect variants in individuals that don’t have an existing classification for the disorder or condition under test. This is both a blessing and curse. The adoption of NGS is widely driven by the potential to improve the diagnostic rate over established PCR or other tests that only look for known and well-categorized biomarkers. Yet, for a small laboratory without a full-time team of dedicated bioinformatic experts and data curators, the burden is left on the variant scientist to do the heavy lifting.
Golden Helix is dedicated to the space of providing fully supported solutions to these small clinical labs. Will the release of VSClinical, we will have a dedicated product focused on each of the major steps of a genetic testing process downstream of the raw data being created by the NGS Sequencing Machine:
- Align & Call: With our offering of Sentieon’s secondary analysis solutions, labs have not only the best performing alignment and small variant calling solution but also the fastest. For many tests, the addition of VS-CNV allows for critical copy number variants to be called without a costly and time-consuming second test
- Annotation & Filtering: VarSeq was designed to be a powerful and flexible tool for designing test specific annotation and filtering workflows, including an unmatched set of bioinformatic algorithms and public as well as licensing annotation sources.
- Variant Interpretation: Now with VSClinical, a dedicated solution allows for the filtered candidate variants to be moved through a guided workflow that reduces subjectivity and improves time-to-classification.
- Reporting: Our powerful and fully customizable clinical reporting solution VSReports allows for the classified and selected variants to be placed into a deliverable that can be passed down to the clinician or another customer of the genetic test.
Standardizing Variant Interpretation
Many early adopters of NGS testing have had to pioneer their own process for variant evaluation. These include some of the larger testing labs including GeneDx, Ambry Genetics and Partners Healthcare (from the top ClinVar submitters list). The need for standardization of the nomenclature used for classification and the scoring process led to the publishing of the ACMG Guidelines paper in 2015. The paper codifies the classification of variants into five levels: Pathogenic, Likely Pathogenic, Variants of Uncertain Significance, Likely Benign and Benign. It also provides 33 scoring criteria that once answered and be combined through a series of rules to arrive at one of these five classifications for a variant. The subsequent adoption of these guidelines in both the US and worldwide demonstrates how badly needed a standardized vocabulary and rubric was needed.
Since that time, several other papers have been published that discuss the adapting of the ACMG guidelines to specific targeted gene tests, laboratories and health networks. VSClinical was built with a comprehensive review of these papers and integrates excerpts and citations at the atomized level of questions and criteria to allow you to deep dive into collective literature to educate your interpretation of the evidence and inform your scoring of each criteria.
Breaking Down Variant Interpretation
When evaluating a variant with an uncertain classification, the lines of evidence used will fall into four categories:
- Population Frequencies: The presence and frequency in large cohorts as an estimation of the maximum tolerated frequency of this variant in humans.
- Gene Function and Variant Impact: Our understanding of how this gene functions, specifically in its relationship to the condition or disorder being tested, as to how the variant impacts that function.
- Clinical Context: How the variant segregates with the family and affected individuals, how it is inherited, and the presence of other variants in the same gene for this individual with an established pathogenicity.
- Studies and Publications: For both the gene impact and function and the clinical case history, the review of published literature and databases is required to get an up to date understanding of the state of our collective knowledge about a variant and the gene it impacts.
We constructed the VSClinical workflow around the concept of grouping the ACMG 33 criteria into four tabs matching these categories. We also put all the summarized evidence and recommendations on the starting tab, allowing the user to deep dive only when necessary into the detailed evidence tabs, while potentially working through variants with decision evidence very quickly and efficiently.
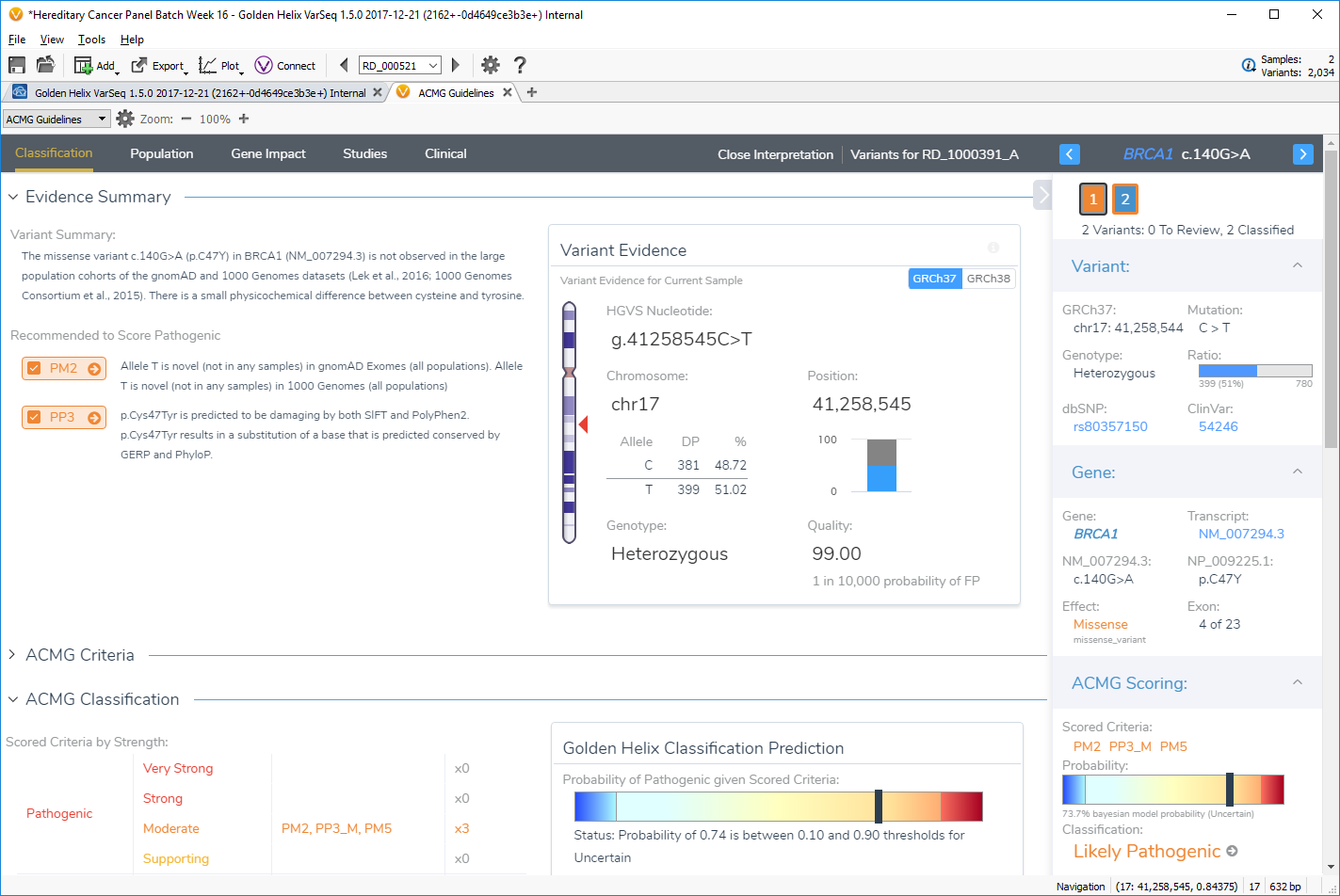
Evidence in VSClinical is available in the four tabs of Population, Gene Impact, Studies, and Clinical. Users start in the Classification tab, where a comprehensive summary of the variant and recommendations is presented along with the ACMG classification and input for the user’s interpretation.
In Part II of this topic, I will go into more detail for each of these tabs discussion the annotation sources and algorithms used to provide supporting evidence and recommendations for the ACMG scoring criteria. Stay tuned!