VSClinical provides a rapid-fire way to investigate any variant’s impact by following the ACMG Guidelines process for classification. We will be demonstrating this by looking at interesting examples of rare disorders and showcasing some evaluation steps users may deploy in their analysis. Our first example in this blog series is for a patient who has an indifference to pain, while all other sensory and motor functions are intact – a disorder known as Congenital Indifference to Pain. Before we dive into VSClinical, I would like to start with a little background on this disorder.
Congenital Indifference to Pain
Congenital indifference to pain (CIP) is an incredibly rare autosomal recessive disorder developed from birth. It was first reported in 1932 in a man who worked as a living pincushion act in the circus (Dearborn GVN. J Nerv Ment Dis. 1932). Interestingly, CIP positive patients register stimuli, have normal joint position sense, vibration, light touch, and temperature perception; they just don’t feel pain. It is critical to differentiate between true CIP and other reduced pain disorders such as hereditary sensory autonomic neuropathies which have abnormal nerve biopsies and abnormal nerve conduction. Loss of function mutations in SCN9A completely cosegregate with this disorder, and while known to be fatal in mice, the loss is not fatal in humans. Also, it has been reported that other voltage-gated sodium channels do not compensate for the lack of Nav1.7 (alternative gene name). Due to the unique impact of losing SCN9A function, it has been considered an ideal target for developing inhibitory drugs for neuropathic and inflammatory pain (Goldberg et al. Clin. Genet 2007). Now that we have some insight into the disorder let’s explore how we can leverage these disorders in a VarSeq workflow.
Example Variant: Target Patient’s Phenotypes
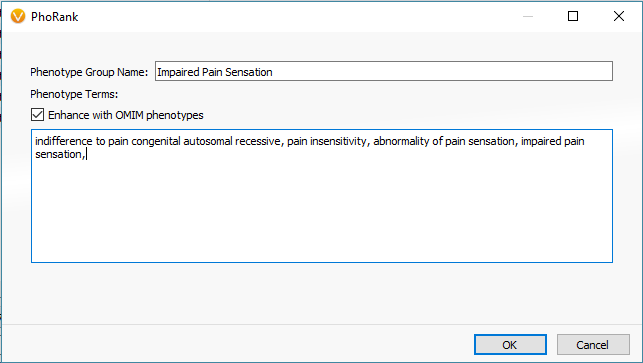
One primary stage of filtering variants is to target those in genes with known associations to the relevant disorders using PhoRank (Figure 1). Mapping across OMIM, Gene Ontology, and Human Phenotype Ontology, PhoRank (Figure 2) outputs a Gene Rank value between 0 – 1 and prioritizes variants with strong associations to the targeted phenotypes (i.e., variants in this sample were filtered for having a Gene Rank value of 0.95 or greater).

Variant Investigation: GenomeBrowse and ACMG Auto Classification
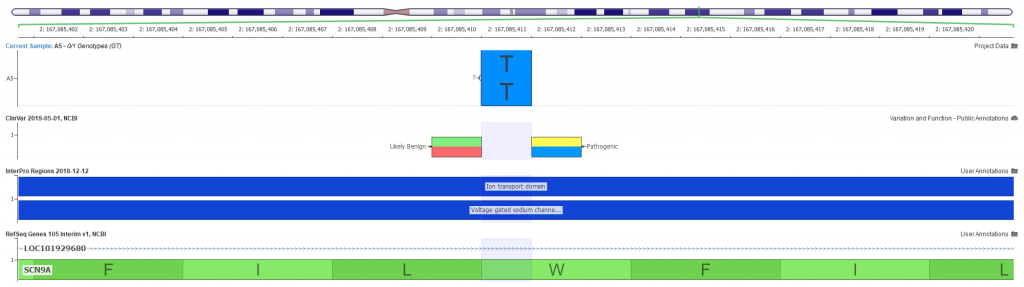
After prioritizing the targeted phenotypes and additional quality filtering, we narrow our search to a homozygous stop gain variant in exon 22 of 27 in the SCN9A gene. Visualizing the variants can be a great way to investigate the landscape of other known problematic variants in the gene. GenomeBrowse is our visualization tool that is integrated into VarSeq made just for this purpose (Figure 3).
Not shown in this figure, but prerun is the auto-classification for this stop gain variant as being likely pathogenic. This auto-classification is the result of running the ACMG classifier algorithm, which considers the summation of all available criteria for each variant. To simplify the collection and evaluation of all ACMG guideline criteria, VSClinical compartmentalizes these criteria into four different sections:
- Population: determining variant frequency
- Gene Impact: variant impact on the gene
- Studies: quick search tool for publications
- Clinical: segregation and inheritance implications for disorder
VSClinical Part I: Variant Population Frequency
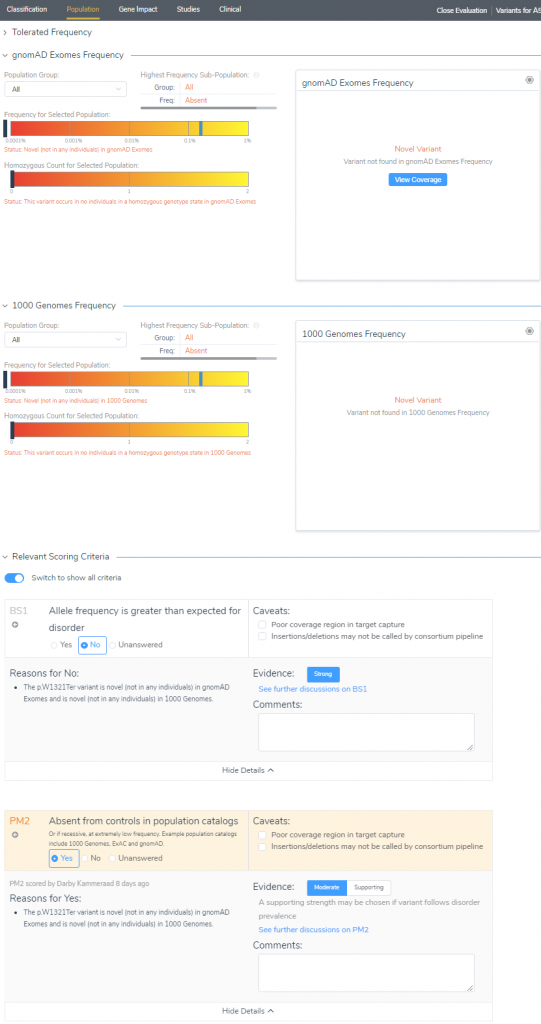
Starting with the Population tab (Figure 4), we see that our SCN9A stop gain is indeed novel for both gnomAD Exomes and 1KGenomes frequency tracks. This makes the assessment of criteria straightforward where we select No for BS1 (allele frequency is greater than expected for the disorder), and Yes to PM2 (absent from controls in population catalogs).
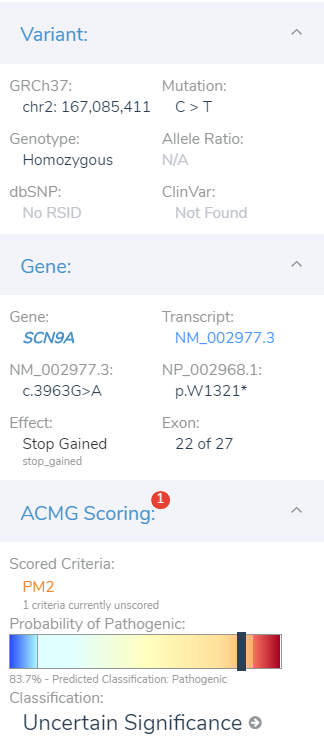
As the criteria are assessed and collected, the variant dashboard will present the current classification criteria as well as the GHI developed classification meter seen in Figure 5. The GHI pathogenicity meter serves as another layer of evidence to help clarify the impact of the variants by feeding all relevant criteria into a probability algorithm. Let’s move on and investigate this variant’s impact on the gene.
VSClinical Part II: Variant Impact on SCN9A
We can get a feel for the impact in the gene by starting with a description of the gene we are in. The SCN9A gene is a voltage-gated sodium channel involved in nociception signaling or pain sensors. The gene impact section of VSClinical provides a lot of great surrounding variant context of what other pathogenic variants exist nearby in this gene. This is shown in Figure 6 below.
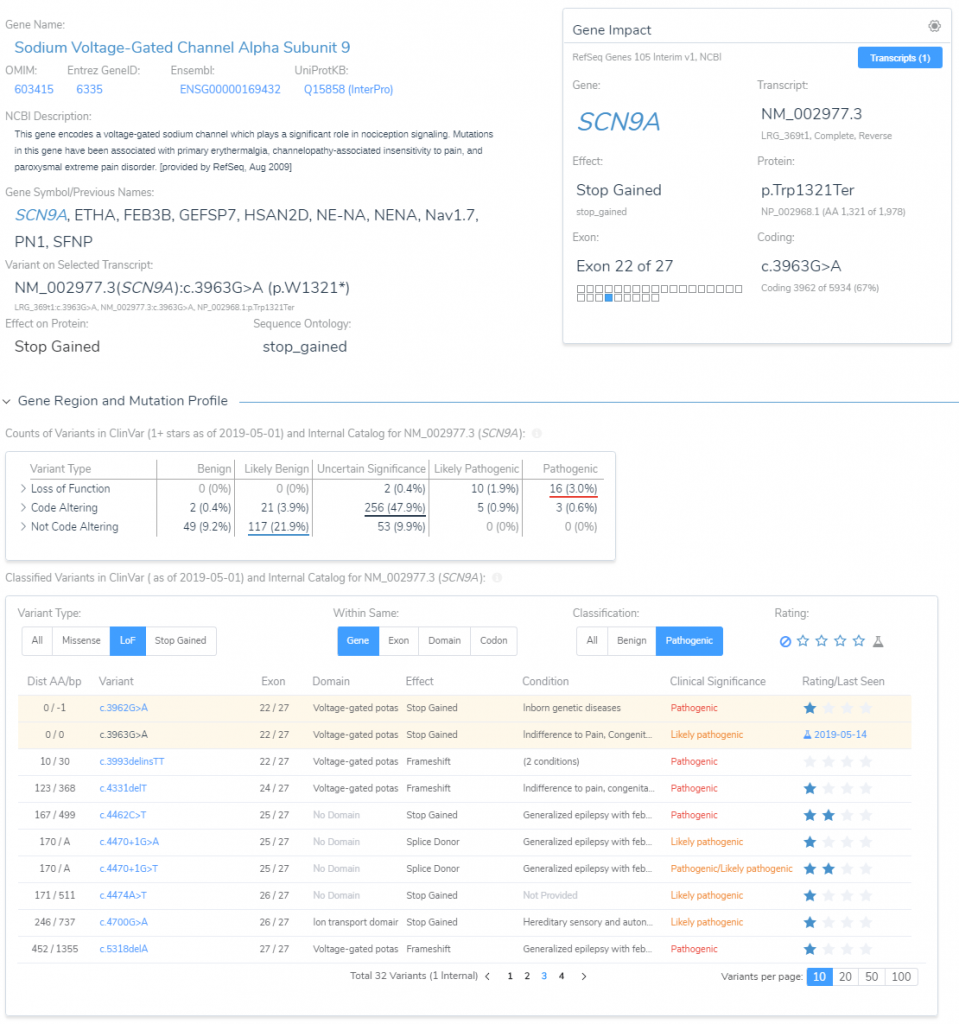
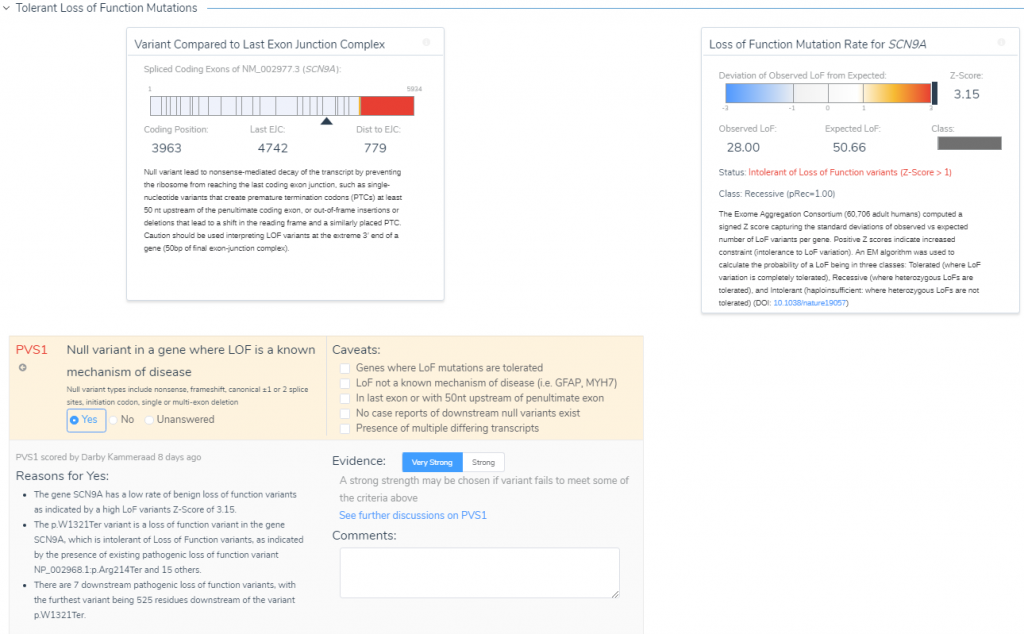
Much of this ‘nearby’ variant context are summarized when reviewing additional criteria like PVS1. Under the Tolerant Loss of Function Mutations section (Figure 7) is the option to review the potential impact of this C>T stop-gain considering the following reasons:
- This gene has a low rate of benign with a high LOF ExAC z-score of 3.15.
- There are other documented pathogenic LOF variants in SCN9A.
- Seven of the documented pathogenic LOF variants are downstream from this variant.
VSClinical Part III: Citing Relevant Literature
Next in line is to assess publications relevant to either the variant under evaluation or any similar nearby variant in the gene. The capture of all relevant literature is done in the Studies section, but before transitioning it is worth noting that the Gene Region and Mutation Profile section of the Gene Impact tab (Figure 8) can help with quickly accessing similar variant type ClinVar submissions and associated literature (Figure 9).
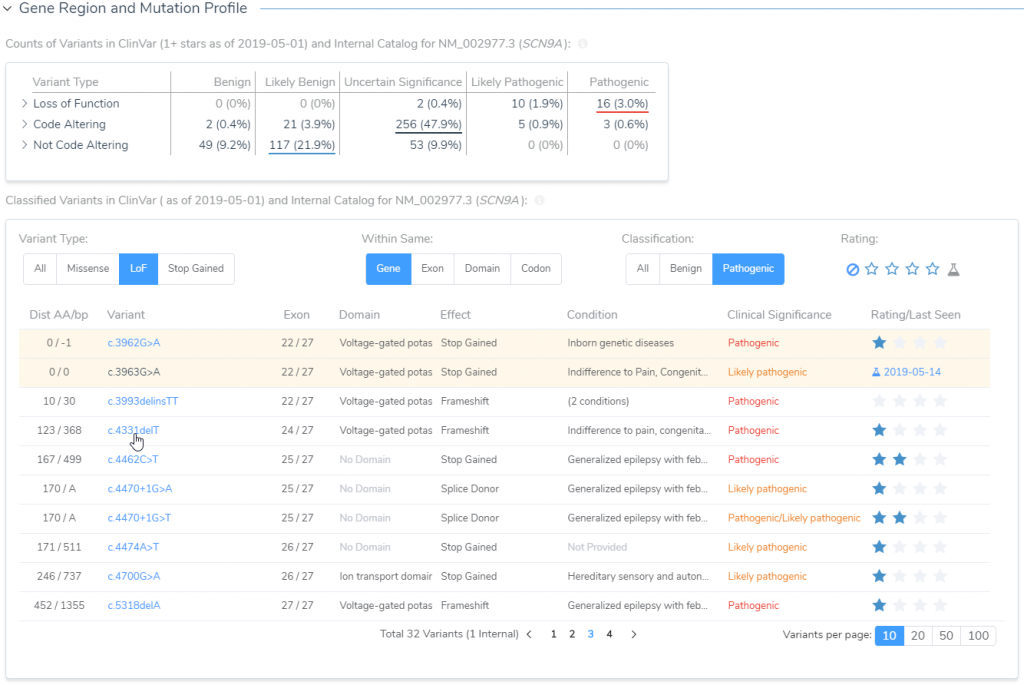
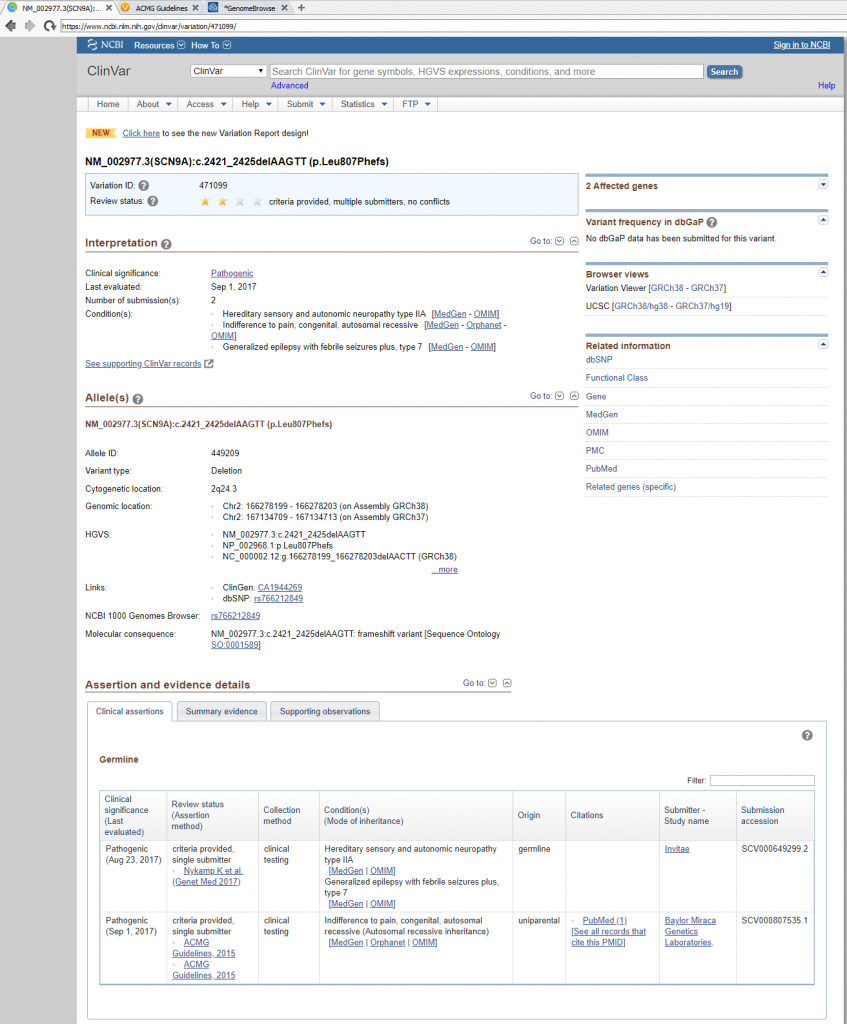
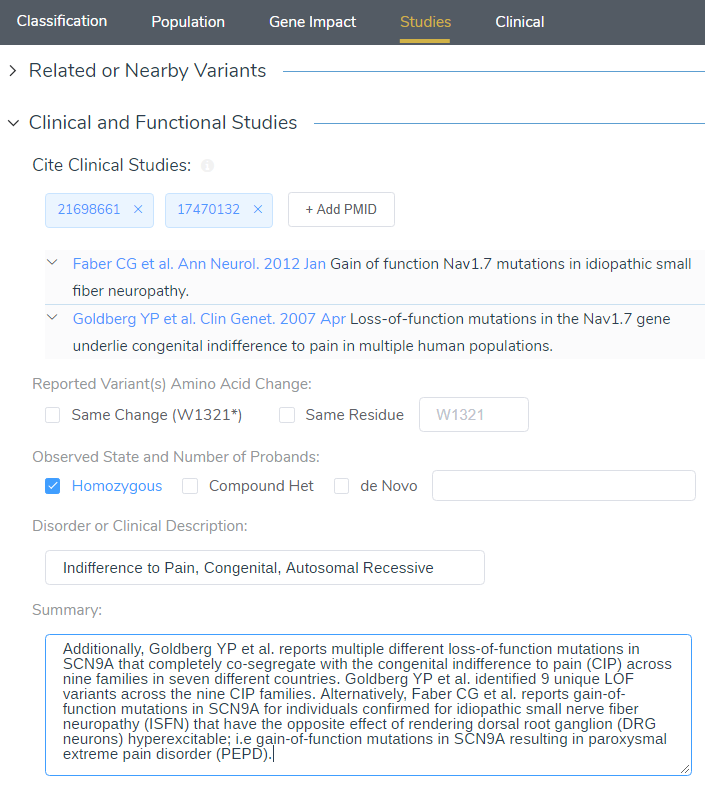
When targeting specific disorders/phenotypes in your VarSeq workflow, the literature search from VSClinical is meant to amplify the auto-interpretation process. Figure 10 demonstrates the capability of directly citing literature via PMIDs and defining the relevant disorders for the patient.
VSClinical Part IV: Final Classification/Interpretation
After browsing through literature and answering all relevant criteria questions, we can get to the final stage of storing our classification and interpretation for the variant. Figure 11 illustrates the final interpretation hub, which can leverage the Auto Interpretation information collected throughout the analysis across each tab and collection of citations from the Studies tab.
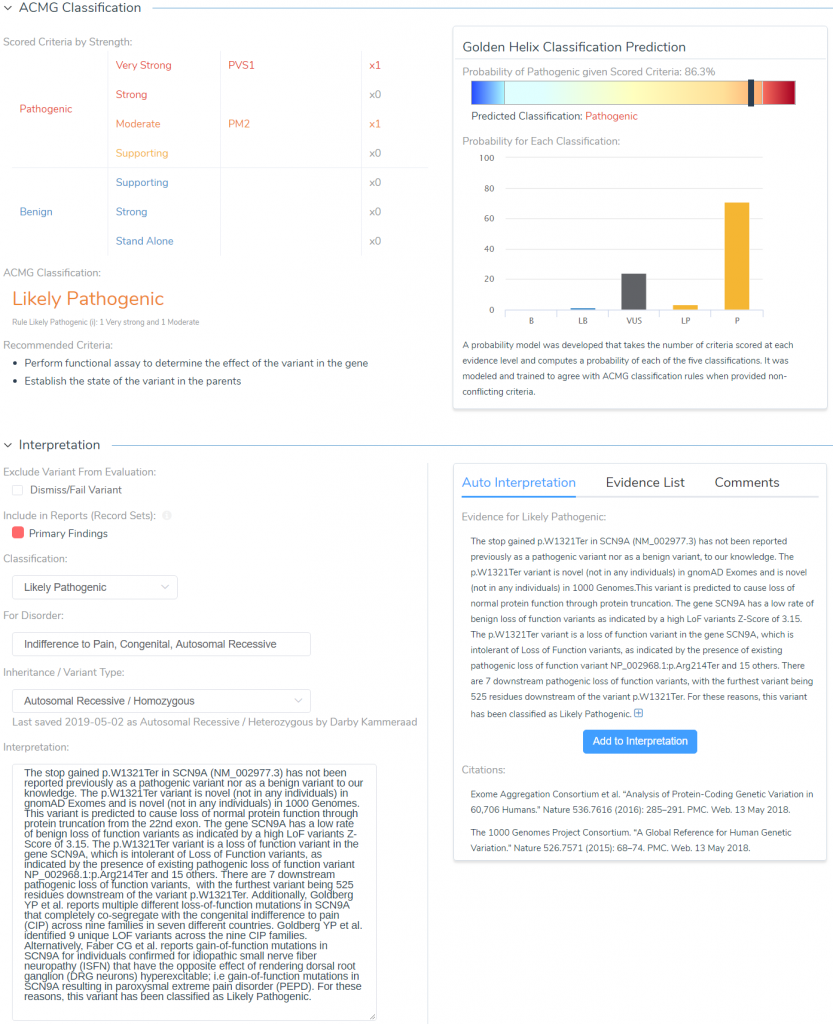
By leveraging the automated collection of all relevant criteria and relevant literature, the final classification and interpretation for our SCN9A stop gain variant are as follows:
The stop gained p.W1321Ter in SCN9A (NM_002977.3) has not been reported previously as a pathogenic variant nor as a benign variant to our knowledge. The p.W1321Ter variant is novel (not in any individuals) in gnomAD Exomes and is novel (not in any individuals) in 1000 Genomes. This variant is predicted to cause loss of normal protein function through protein truncation from the 22nd exon. The gene SCN9A has a low rate of benign loss of function variants as indicated by a high LoF variants Z-Score of 3.15. The p.W1321Ter variant is a loss of function variant in the gene SCN9A, which is intolerant of Loss of Function variants, as indicated by the presence of existing pathogenic loss of function variant NP_002968.1:p.Arg214Ter and 15 others. There are 7 downstream pathogenic loss of function variants, with the furthest variant being 525 residues downstream of the variant p.W1321Ter.
Additionally, Goldberg YP et al. reports multiple different loss of function mutations in SCN9A that completely co-segregate with the congenital indifference to pain (CIP) across nine families in seven different countries. Goldberg YP et al. identified 9 unique LOF variants across the nine CIP families. Alternatively, Faber CG et al. reports gain-of-function mutations in SCN9A for individuals confirmed for idiopathic small nerve fiber neuropathy (ISFN) have the opposite effect of rendering dorsal root ganglion (DRG neurons) hyperexcitable; i.e. gain-of-function mutations in SCN9A resulting in paroxysmal extreme pain disorder (PEPD). For these reasons, this variant has been classified as Likely Pathogenic.
The goal for this blog is to provide a simple presentation on the profound usability of VSClinical to assist with consistent and comprehensive variant classification and interpretation. Additionally, this blog is meant to showcase the software capabilities and does not constitute a final clinical assessment from healthcare professionals. As mentioned, this is the first of many example variant blog posts to come where future examples will explore other variant effects and other interesting disorders. Each sample or patient has unique conditions, and if you would like guidance that is specific to your use case please reach out to support@goldenhelix.com for more formal training.

Check out some of the other example variants in this series:
- Variant Interpretation with VSClinical: Evaluation of an X-linked recessive mutation
- Variant Interpretation with VSClinical: Evaluation of Hypertrophic Cardiomyopathy
- Variant Interpretation with VSClinical: Congenital Myasthenic Syndromes (CMS)
- Variant Interpretation with VSClinical: Huntington’s Disease (HD)
- Variant Interpretation with VSClinical: Non-Small Cell Lung Cancer
- Variant Interpretation with VSClinical: RET-KIF5b Gene Fusion
Citations:
- Goldberg YP, et al. Loss-of-function mutations in the Nav1.7 gene underlie congenital indifference to pain in multiple human populations. Clin Genet 2007: 71: 311-319.
- Faber CG, et al. Gain of Function Nav1.7 Mutations in Idiopathic Small Fiber Neuropathy. Ann Neurol. 2012: 71: 26-39.
- Dearborn GVN. A case of congenital pure analgesia. J Nerv Ment Dis 1932: 75: 612-615.