In a recent webcast, our VP of Product and Engineering Gabe Rudy gave us insight into the current capability and benefits to lifting over to the GRCh38 assembly. Golden Helix fully supports this transition into the most recent reference assembly and have developed our tools on both the 38 and 37 fronts. The purpose of this blog is to not only illustrate the value of
SRGAP2 is a great example of one such troublesome region. First, let us look at the differences in the SRGAP2 region between 37 and 38 (Figures1 & 2). Even at a broad glance, you notice the transcripts-enriched 38 based SRGAP2 and if you were to zoom in you would find it also lists UTR regions which are missing from 37. Differences exist even outside of SRGAP2 in neighboring regions/genes. Tracking all the finer details of these differences may prove to be tedious; which is why we have taken the liberty of curating some helpful annotations that can easily define some patched-up regions.
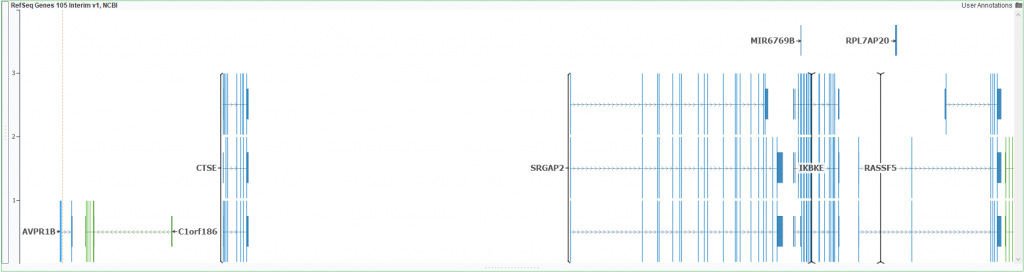
Figure 1: 37 based assembly and RefSeq genes plotted for SRGAP2 and surrounding regions
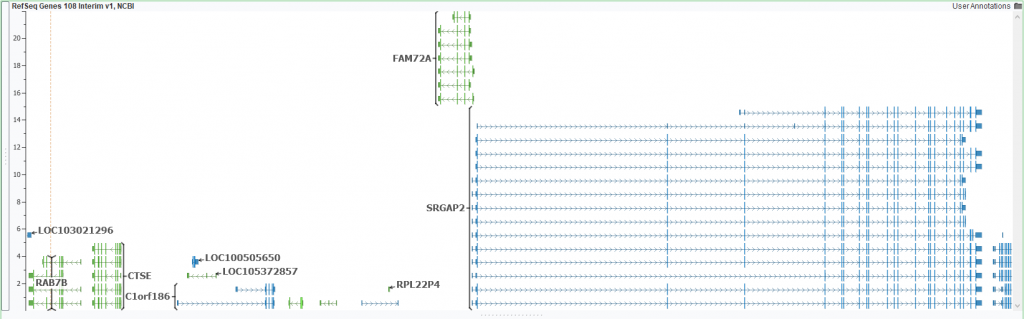
Figure 2: 38 based assembly and RefSeq genes plotted forSRGAP2
In version 2.1.0, we’ve added new annotations that
- Contigs Dropped or Changed from GRCh37 to GRCh38
- Patchesto GRCh37 Reference Sequence
If you wish to maintain using 37, you may want to consider incorporating these annotations into your filter chain/genome browse view to ensure you are accounting changed/dropped regions in the recent 38
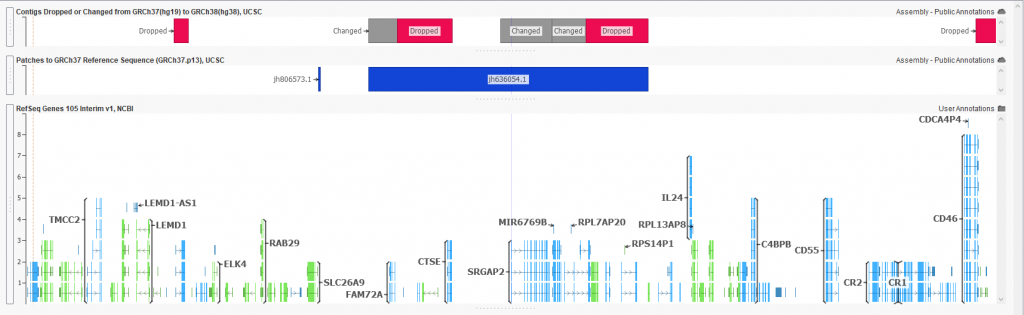
Figure 3: New annotations highlighting dropped/changed regions and
fixes made in the new 38 based assembly.
Also, it is worth investigating the overlapping “patched” regions to see the fixes made in 38 (Figure 4).
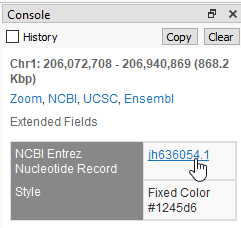

Figure 4: NCBI hyperlink for details on patches over SRGAP2 and neighboring regions in the 37 reference assembly
The fundamental reason as to why this is so important is best demonstrated with some SRGAP2 variants being analyzed in the variant table. Under theHGVS p. notation in RefSeq, you can see that the 37 based p.notation is missing (i.e.,p.?)(Figure 5). Not only do we capture the full p.notation in 38 (Figure 6) by incorporating the patches, but you also account for any possible impact this may have on ACMG classification with VSClinical.
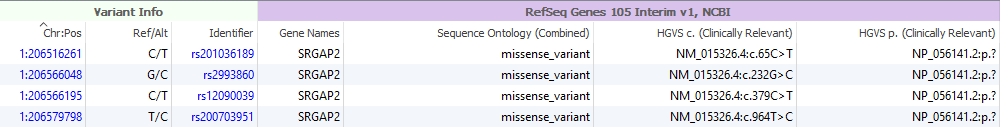
Figure 5: Missing p. notation with the 37-based project
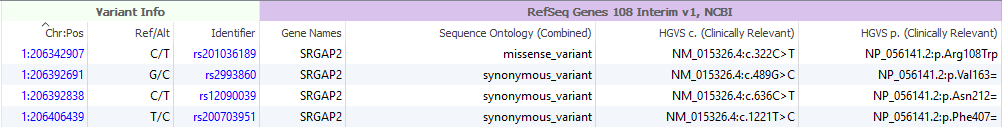
Figure 6: After liftover, the p.notation is present in 38
We will continue to support our GRCh37-based users through feature/annotation development but also wanted to encourage the transition to 38. Making the switch can help users save a lot of hassle when dealing with troublesome genes like SRGAP2, but also could be considered a best-practice by using the highest quality reference assembly to date. If you have any additional questions on the justifications of switching to