Compound Heterozygous Workflows: Including a 2nd Affected Child
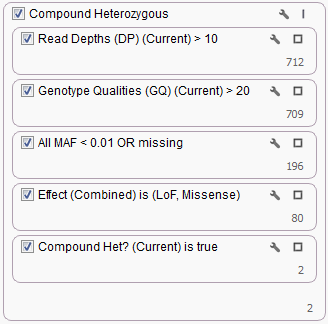
Looking for Compound Heterozygous regions for a trio is fairly straight forward in VarSeq, we include this workflow in our shipped Exome Trio Template. An example of which is included with our Example Projects which can be found by going to File > Example Projects > Example YRI Exome Trio Analysis.
But what if instead of a Trio you had data for a Quad (mother, father and 2 affected children), how would you incorporate this additional information into your Compound Heterozygous workflow? You could of course treat them as two separate Trios and use the existing workflows to complete your analysis. But what if you only wanted to identify Compound Heterozygous Regions that were in common between the two children. The existing workflow can be easily customized by duplicating filter cards and changing sample options to pull data from specific samples.
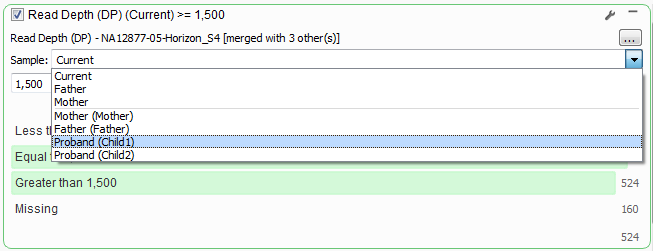
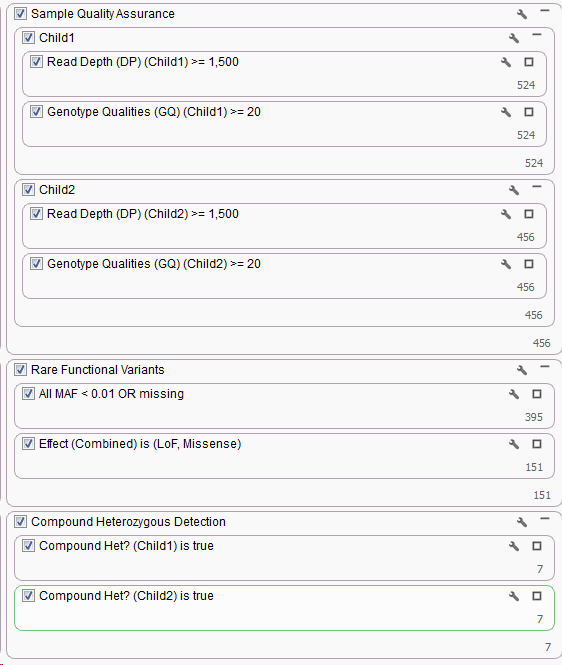
The first two cards listed in the above filter chain for Read Depth and Genotype Qualities are only looking at the “Current” sample, in terms of a Trio that means the first Proband that was imported. If you click the wrench icon in the upper right corner of the Read Depth filter card you can switch the Sample information to always pull the Read Depth from the first affected child instead.
Then duplicate the card by right-clicking and selecting Duplicate and update the card to pull from the second affected child. Repeat the process for the Genotype Qualities filter card. Adding a few Filter Containers the resulting quality control part of the filter chain would look something like the following.
The next two filter cards for Minor Allele Frequency and Variant Transcript Effect would not change with the addition of another sample as they are variant level filters, so creating a Filter Container and dragging them into it would look like the following.
Now to look at the Compound Heterozygous output for each child. Similar to the changes for the Read Depth filter card above, switch the Sample options to Child1 then duplicate the card and specify Child2.
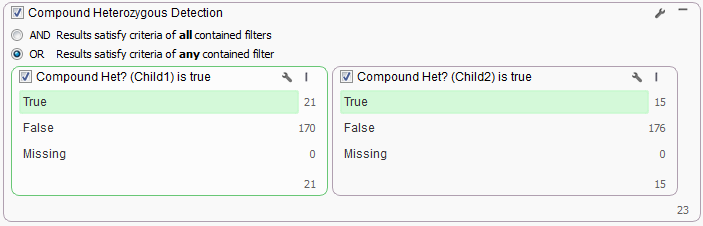
To look at the Compound Het regions for each child separately create a Filter Container for the Compound Het cards and set the container criteria as “OR” by clicking the wrench icon in the upper right corner of the container.
At some point you will be prompted to re-run the Compound Het algorithm once the changes to the cards are made as the algorithm is run on the filtered data set not on all variant originally imported.
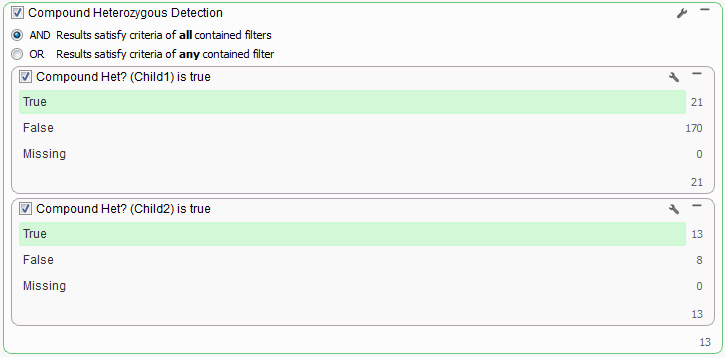
For the data used for this example workflow, Child1 has 21 variants that make up 6 Compound Het genes (to view these genes in your project click on the number at the bottom of the left card and open the Variants by Gene table) and Child2 has 15 variants that make up 4 Compound Het gene regions. With this data set it is fairly easy to visually inspect the results of each child to pick out regions in common between the two children, however if you are dealing with whole exome sequencing data the possible Compound Hets for each child could be in the hundreds. To narrow down the list to only those they have in common is a simple as changing the Filter Container criteria to “AND” at the top of the card.
Combining all the above changes into one Filter Chain and we have our Quad workflow.