As the need to educate prospective healthcare professionals in the interpretation of genetic data increases, Dr. Jeffrey Moore at the University of Illinois – Urbana-Champaign is using genetics in his chemistry courses. In doing so, Moore is creating a strong connection between the content in his courses and the underlying principles of health and medicine.
Last year, Dr. Moore presented a webcast (The Molecular Sciences Made Personal) to the community. This webcast discussed the use of his students’ personal genetic data to promote curiosity-driven learning.
This year, Moore’s Introduction to Chemistry class used Pharmacogenomics to strengthen the principles of kinetics and thermodynamics of metabolism. Over the semester, students in Moore’s class used the genomic data of Billy Ashcraft from Harvard’s Personal Genome Project to determine Billy’s drug response based on his genetic variants.
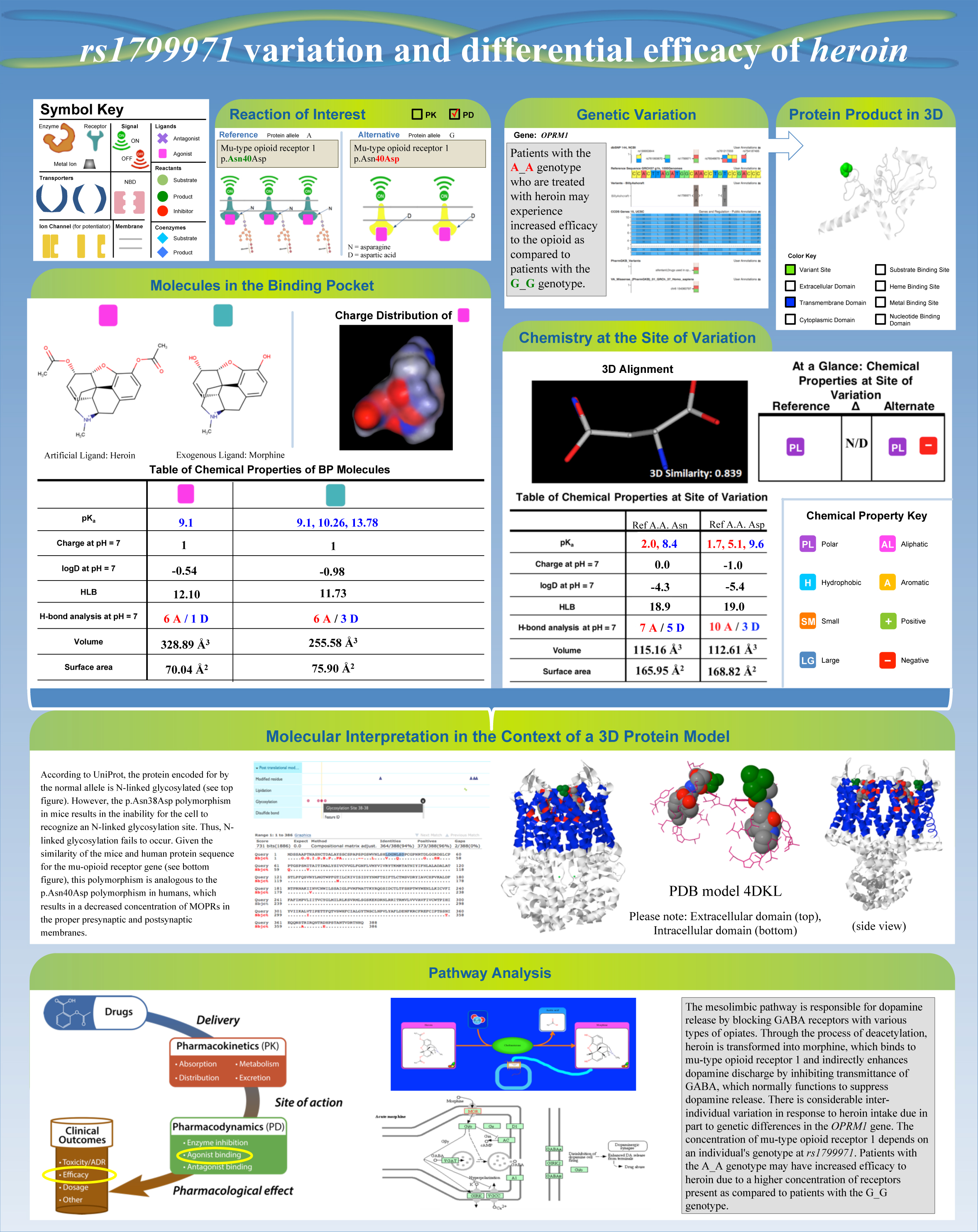
A final poster presented by a group of student’s in Dr. Moore’s CHEM 104 class
Dr. Moore’s students used Golden Helix’s GenomeBrowse to investigate possible genetic variations affecting drug efficacy. To do so, the students downloaded the PharmGKB database file with genomic coordinates and created a track of missense variants in GenomeBrowse to find the regions affected.
Dr. Moore is having great success using Golden Helix’s suite of software products in his classroom in order to strengthen the relationship between genetics and organic chemistry. He is getting great reviews from his students as well.
If you are interested in learning more about using our software tools in your curriculum, please email us at info@goldenhelix.com.