Even though GRCh37 is currently the most widely used human genome assembly, GRCh38 provides a more complete human reference genome, offers more accurate genomic analysis, and includes centromere and mitochondrial information.
However, we’re getting ahead of ourselves. Perhaps start with how we got here.
The Human Genome Project started this all off with the world’s largest biological collaboration project in 1990 (although talks begin back in the mid-1980s). Over a decade later it was declared done in 2003, although further work was being conducted.
The next big milestone was the release of NCBI36 in 2006. This assembly, which is also referred to as the hg18 assembly by the University of California Santa Cruz (UCSC), was the first genome assembly employed with the task of aligning reads from high-throughput sequencers (the Illumina GAII) and was used by the pilot project for 1000 Genomes to call millions of variants.
Three years later, the Genome Reference Consortium (GRCh) gives us what is currently the most widely used GRCh37 (or hg19) human genome assembly. In a watershed moment that helped cement its use in genomics, the 1000 Genomes Project switched all analysis to GRCh37, and in the era of the Illumina HiSeq 2000 sequencing system, many labs started using it for their NGS analysis.
Although it seems like a recent release, the new and improved GRCh38 (hg38) track was published back in 2013 with more sequences, centromere representation, and the mitochondrial reference sequence. GRCh38 improved upon GRCh37 by correcting erroneous bases and misassembled regions and correcting over 100 assembly gaps.
- Alternate sequences
The GRCh38 track more adequately acknowledges the high amount of variability that can arise in specific regions preventing enough representation by a single sequence by providing alternate sequences, or alternative loci. Providing over 250 alternative loci in GRCh38 touches over 150 genes; 25% of which are medically interpretable.
- Centromere
Models created by the combined efforts of Duke University and UCSC now help the mapping of centromeric regions that were previously represented as large gaps.
- Mitochondrial
This little 16+ kilobase sequence is included in the GRCh38 assembly and is the Revised Cambridge Reference Sequence (rCRS) from MITOMAP. In encodes 37 genes and displays a slightly different genetic code than nuclear genes.
Some readers at this point may be asking, “Does the switch to GRCh38 make a difference?” Moreover, the answer is yes. The GRCh38 assembly displays better alignment, fewer “frame-fixing” introns, better gene representation, and more annotation sources are using GRCh38 natively. ICGC, COSMIC, and TopMed have switched over to name a few.
An example of a better gene representation is shown below in the EMG1 gene where the top picture shows a frame-fixing intron on the right side of the gene in the GRCh37 mapping, but this intron is not needed in the GRCh38 assembly below it.
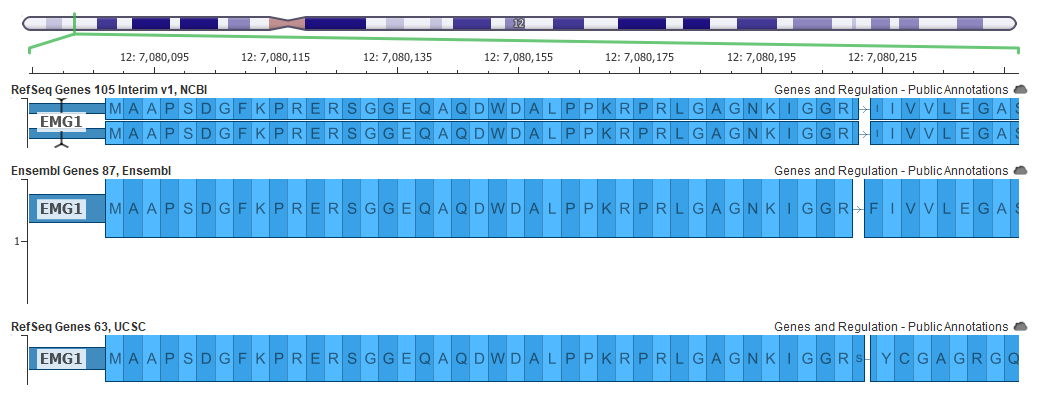
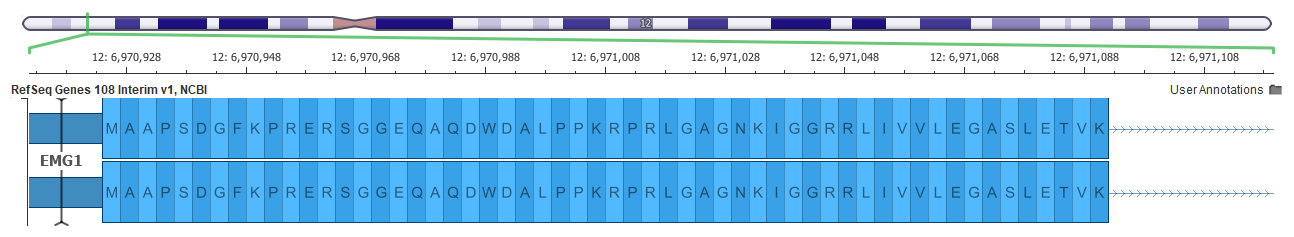
Now all these changes can affect variant interpretation. The main tool within the Golden Helix suite for variant interpretation within the ACMG guidelines is VSClinical.

VSClinical allows for easy, intuitive, and repeatable variant analyses within GRCh37 and GRCh38 assembly frameworks. For example, the image below shows a toggle switch in the upper right-hand side of the Variant Evidence card that allows the user to easily switch between the two assemblies for variant representation.
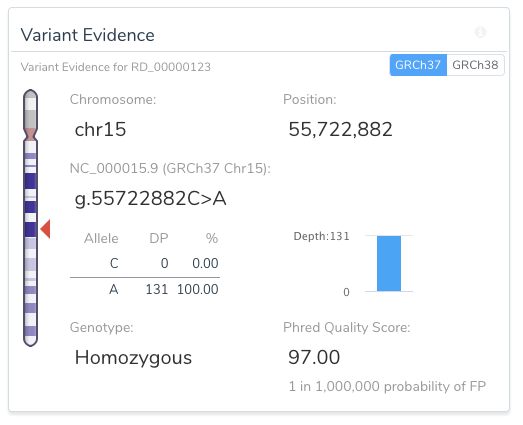
Another example of the ease in which VSClinical increases informs users of unique variant traits is the variant shown as a pure reference artifact in GRCh37 (the insertion of the G allele).
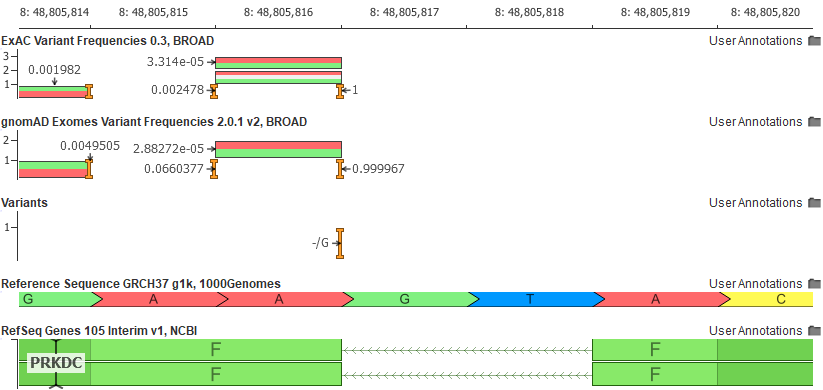
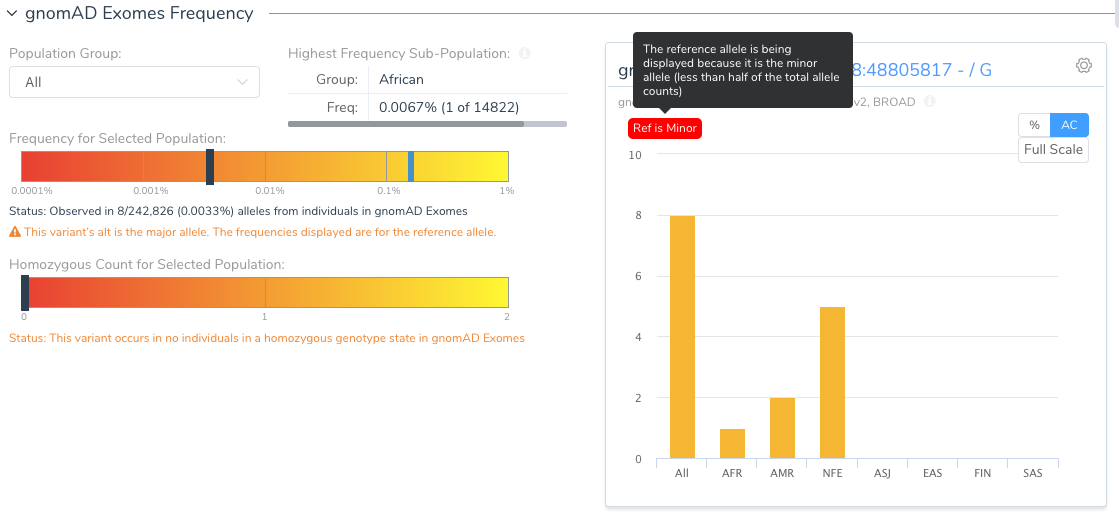
The Population tab in the VSClinical analysis will flag this variant and show the user that the reference allele is, in fact, the minor allele.
Why would the reference allele be so rare? Remember the reference genome is still largely a composite of the genomes of 13 anonymous volunteers from the Buffalo, New York area and any given segment of the reference genome is attributed to a single individual. As we have now thousands of samples to compare to, we can find instances where the haplotype of the reference harbors extremely rare alleles. The GRCh38 reference has “fixed” a number of these rare letters, especially in cases where the rare allele encoded a rare protein change.
So the next question may be, “How can I switch over to GRCh38 if all of my current data uses GRCh37?” Well, for any labs looking to incorporate GRCh38, VarSeq and VSClinical are not only including more GRCh38 annotation tracks but allow for easily “lifting over” any GRCh37 tracks to GRCh38. The example below shows how a user can LiftOver sample VCF files from GRCh37 to GRCh38 on import.
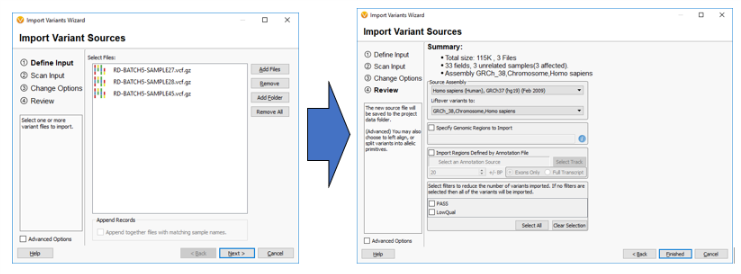
It should also be noted that this software allows for the reverse to happen and GRCh38 tracks and sample data can be lifted over to GRCh37 if needed.
If you have any questions or comments or would like to start migrating over to GRCh38 in your research, please contact Golden Helix here. Moreover, to view the full webcast, please do so here.