Relating human phenotypes to genotypes is the name of the game with OMIM, and as their website says, “is intended for use primarily by physicians and other professionals concerned with genetic disorders, by genetics researchers, and by advanced students in science and medicine.” The Online Mendelian Inheritance in Man (or OMIM) was originally created by Dr. Victor A. McKusick in the 60s as a catalog called the Mendelian Inheritance in Man (MIM), but the ‘O’ was added in 1985 when the catalog was put online with the help of the Johns Hopkins University School of Medicine which still edits and adds to the catalog today. Users should keep in mind, though, that is still common to see both MIM and OMIM used in references. OMIM has gained the reputation of containing genetic information about not only common genotypical and phenotypical relationships, but many lesser-known and edge-case relationships, as well as the clinical citations linked to individual genetic mutations, clinical citations and various other lines of evidence useful for interpreting human genetics. Although the collection is available to the public, easily using the information for annotation, analysis, or diagnosis requires much more effort than hitting the download button (which only exists for academic users).
Annotating OMIM
The OMIM catalog is updated daily and Golden Helix updates its OMIM snapshot annotations on a monthly basis to include Gene Annotations, Phenotype Annotations (linked to genes), Variant Annotations, and Region Annotations. The image below shows the OMIM track options for annotation in VarSeq. Both the Gene and Phenotype tracks offer an additional option of including details (or, “with Details”). The track with details offers, as the name implies, more information on each annotation site such as Clinical Features, History, and Cytogenetics. As a result, these tracks including details are about 8 times larger than the version without.
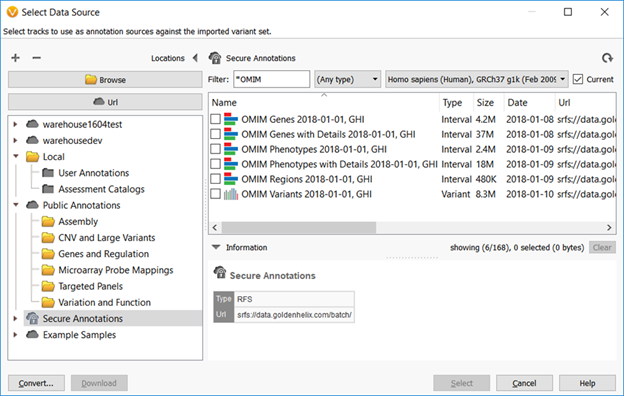
To extract this variety of tracks from the OMIM data source, data curation involves the use of additional parsing tools. Correlating the genomic location and variant information is performed using the most current versions of dbSNP and RefSeq. These tools allow extensive searches in the OMIM database to neatly split out the corresponding gene, variant, or region-based tracks. By this description, both the Gene Track and Phenotype Track fall under the gene-based track designation where the Phenotype Track provides one phenotype record per gene. This is incredibly useful for inheritance and phenotype analysis where a gene name, a phenotype description, or a genomic location is the start of a search. The newest OMIM track type to join the roster is the OMIM Regions track.
OMIM Regions Track
Golden Helix has offered OMIM genes and phenotypes annotation tracks but now offers an OMIM regions track. While many OMIM entries are specific to a gene, some are more broadly defined as diseases associated to genomic regions and sometimes to specific copy number states of that region (Gains vs Losses). This track is especially useful with CNV analysis in both VarSeq and SVS.
The GenomeBrowse plot for an annotation is shown below with the OMIM Regions, OMIM Phenotypes with Details, OMIM Variants, and OMIM Genes with Details above the RefSeq genomic location.
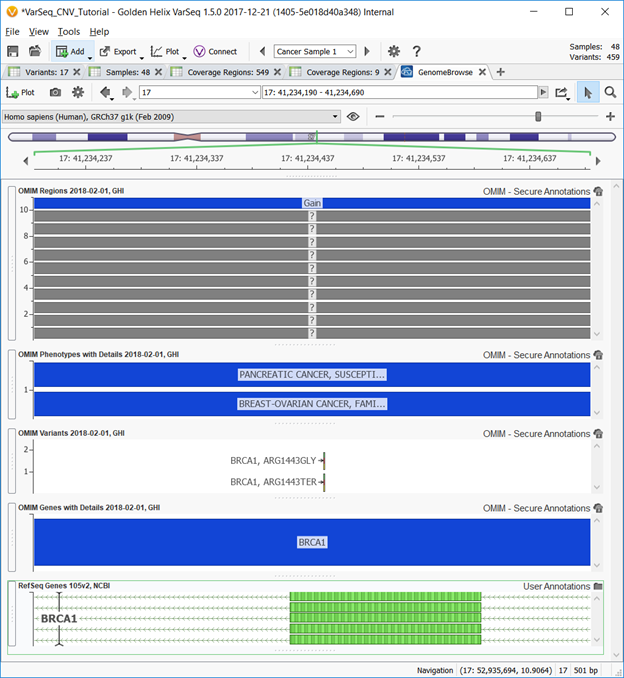
The OMIM Stack
Altogether, the OMIM annotation stack includes tracks based on a Gene, Phenotype, Variant and Regional basis. Golden Helix offers these for both GRCh37 and GRCh38 reference sequences bringing the total OMIM stack offering (including the “with Details”) options up to 12. Curation of the OMIM catalog allows users to determine the variety of phenotypes linked to certain genes and genomic regions, the function of these phenotypes, and variant interpretations all with specific paper and PubMed references. OMIM does an amazing job of collecting records and citations of human genetic and
phenotypic data and Golden Helix sifts and sorts the lists into a variety of easily useable annotation tracks. Please contact us if you are interested in adding this feature to your existing license.