Dr. Bryce Christensen recently gave a webcast on Maximizing Public Data Sources for Sequencing and GWAS Studies in which he covered options for getting GWAS and sequence information online, tips for working with these datasets and what you’ll see in terms of data quality and usefulness, how to use public data sources in conjunction with your GWAS or sequence study… Read more »
Utilizing Identical Twins Discordant for Schizophrenia to Uncover de novo Mutations We are living in exciting times – the reality of high-resolution Cand individual genome sequencing now offers renewed hope in the search for the causes of complex diseases. When this technology is combined with genetic relationships, individual sequences add unrivaled proficiency. Our lab is located in London, Ontario, Canada… Read more »
Thanks to everyone for the great webcast yesterday. We had over 850 people register for the event and actually broke the record! Take that Bryce and Gabe! If you would like to see the recording, view it at: Mixed Models: How to Effectively Account for Inbreeding and Population Structure in GWAS. While preparing for this webcast, we chose to focus… Read more »
Presenter: Greta Linse Peterson, Senior Statistician Date: Wednesday, June 5th, 2013 Time: 12:00 pm EDT, 60 minutes Abstract Population structure and inbreeding can confound results from a standard genome-wide association test. Accounting for the random effect of relatedness can lead to lower false discovery rates and identify the causative markers without over-correcting and dampening the true signal. This presentation will… Read more »
A few months ago, our CEO, Christophe Lambert, directed me toward an interesting commentary published in Nature Reviews Genetics by authors Bjarni J. Vilhjalmsson and Magnus Nordborg. Population structure is frequently cited as a major source of confounding in GWAS, but the authors of the article suggest that the problems often blamed on population structure actually result from the environment… Read more »
Last week Khanh-Nhat Tran-Viet, Manager/Research Analyst II at Duke University, presented the webcast: Insights: Identification of Candidate Variants using Exome Data in Ophthalmic Genetics. (That link has the recording if you are interested in viewing.) In it, Khanh-Nhat highlighted tools available in SVS that might be under used or were recently updated. These tools were used in his last three… Read more »
In preparation for a webcast I’ll be giving on Wednesday on my own exome, I’ve been spending more time with variant callers and the myriad of false-positives one has to wade through to get to interesting, or potentially significant, variants. So recently, I was happy to see a message in my inbox from the 23andMe exome team saying they had… Read more »
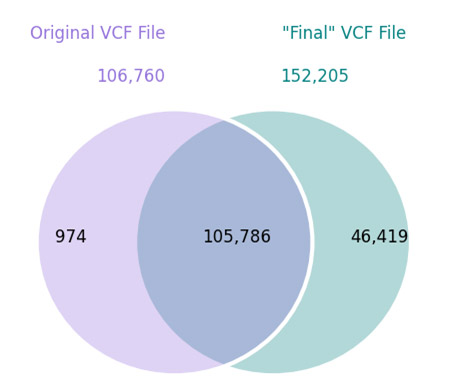
After my latest blog post, Jeffrey Rosenfeld reached out to me. Jeff is a member of the Analysis Group of the 1000 Genomes Project and shared some fascinating insights that I got permission to share here: Hi Gabe, I saw your great blog post about the problems in the lack of overlap between Complete Genomics and 1000 Genomes data. I… Read more »
Thank you to everyone who joined us yesterday for a webcast by Dr. Ken Kaufman of Cincinnati Children’s Hospital: “Identification of Candidate Functional Polymorphism Using Trio Family Whole Exome DNA Data.” Over 750 people registered for this event and 430 attended – a new Golden Helix record! If you missed the webcast (or would like to watch it again), the… Read more »
I recently curated the latest population frequency catalog from the 1000 Genomes Project onto our annotation servers, and I had very high hopes for this track. First of all, I applaud 1000 Genomes for the amount of effort they have put in to providing the community with the largest set of high-quality whole genome controls available. My high hopes are… Read more »
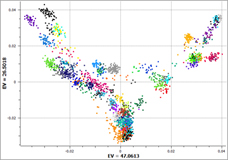
My work in the GHI analytical services department gives me the opportunity to handle data from a variety of sources. I have learned over time that every genotyping platform has its own personality. Every time we get data from a new chip, I tend to learn something new about the quirks of genotyping technology. I usually discover these quirks the… Read more »
As I’ve mentioned in previous blog posts, one of the great aspects of our scientific community is the sharing of public data. With a mission of providing powerful and accurate tools to researchers, we at at Golden Helix especially appreciate the value of having rich and extensive public data to test and calibrate those tools. Public data allow us to… Read more »
The Golden Helix sales team recently came to me for recommendations regarding best practices for incorporating public controls in SNP GWAS. It seems that there has been a surge of questions regarding this practice over the past few weeks from our customers. Initially, I laughed at the irony of being asked to outline the best practices for what I see… Read more »
Over the past 3 years, Golden Helix has analyzed dozens of public and customer whole-genome and candidate gene datasets for a host of studies. Though genetic research certainly has a number of complexities and challenges, the number one problem we encounter, which also has the greatest repercussions, is born of problematic experimental design. In fact, about 95% of the studies… Read more »
In the paper Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia, Todd Lencz, Ph.D. introduced a new way of doing association testing using SNP microarray platforms. The method, which he termed “whole genome homozygosity association”, first identifies patterned clusters of SNPs demonstrating extended homozygosity (runs of homozygosity or “ROHs”) and then employs both genome-wide and regionally-specific statistical tests… Read more »